[English] 日本語
 Yorodumi
Yorodumi- EMDB-29281: Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | |||||||||||||||
 Map data Map data | Map of STING tetramer bound to cGAMP and NVS-STG2 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | STING / innate immunity / molecular glue / IMMUNE SYSTEM | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationSTING complex / STAT6-mediated induction of chemokines / protein localization to endoplasmic reticulum / 2',3'-cyclic GMP-AMP binding / cyclic-di-GMP binding / STING mediated induction of host immune responses / serine/threonine protein kinase complex / positive regulation of type I interferon-mediated signaling pathway / IRF3-mediated induction of type I IFN / cGAS/STING signaling pathway ...STING complex / STAT6-mediated induction of chemokines / protein localization to endoplasmic reticulum / 2',3'-cyclic GMP-AMP binding / cyclic-di-GMP binding / STING mediated induction of host immune responses / serine/threonine protein kinase complex / positive regulation of type I interferon-mediated signaling pathway / IRF3-mediated induction of type I IFN / cGAS/STING signaling pathway / proton channel activity / reticulophagy / pattern recognition receptor signaling pathway / cytoplasmic pattern recognition receptor signaling pathway / cellular response to exogenous dsRNA / protein complex oligomerization / autophagosome membrane / positive regulation of macroautophagy / autophagosome assembly / : / cellular response to interferon-beta / positive regulation of type I interferon production / positive regulation of defense response to virus by host / endoplasmic reticulum-Golgi intermediate compartment membrane / signaling adaptor activity / antiviral innate immune response / activation of innate immune response / autophagosome / protein serine/threonine kinase binding / positive regulation of interferon-beta production / secretory granule membrane / Regulation of innate immune responses to cytosolic DNA / cytoplasmic vesicle membrane / SARS-CoV-1 activates/modulates innate immune responses / peroxisome / regulation of inflammatory response / defense response to virus / DNA-binding transcription factor binding / RNA polymerase II-specific DNA-binding transcription factor binding / mitochondrial outer membrane / endosome / cilium / ciliary basal body / Golgi membrane / innate immune response / ubiquitin protein ligase binding / Neutrophil degranulation / protein kinase binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm / SARS-CoV-2 activates/modulates innate and adaptive immune responses / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
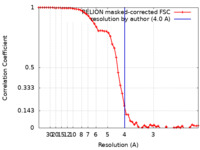
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Li J / Canham SM / Zhang X / Bai X / Feng Y | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Activation of human STING by a molecular glue-like compound. Authors: Jie Li / Stephen M Canham / Hua Wu / Martin Henault / Lihao Chen / Guoxun Liu / Yu Chen / Gary Yu / Howard R Miller / Viktor Hornak / Scott M Brittain / Gregory A Michaud / Antonin Tutter / ...Authors: Jie Li / Stephen M Canham / Hua Wu / Martin Henault / Lihao Chen / Guoxun Liu / Yu Chen / Gary Yu / Howard R Miller / Viktor Hornak / Scott M Brittain / Gregory A Michaud / Antonin Tutter / Wendy Broom / Mary Ellen Digan / Sarah M McWhirter / Kelsey E Sivick / Helen T Pham / Christine H Chen / George S Tria / Jeffery M McKenna / Markus Schirle / Xiaohong Mao / Thomas B Nicholson / Yuan Wang / Jeremy L Jenkins / Rishi K Jain / John A Tallarico / Sejal J Patel / Lianxing Zheng / Nathan T Ross / Charles Y Cho / Xuewu Zhang / Xiao-Chen Bai / Yan Feng /  Abstract: Stimulator of interferon genes (STING) is a dimeric transmembrane adapter protein that plays a key role in the human innate immune response to infection and has been therapeutically exploited for its ...Stimulator of interferon genes (STING) is a dimeric transmembrane adapter protein that plays a key role in the human innate immune response to infection and has been therapeutically exploited for its antitumor activity. The activation of STING requires its high-order oligomerization, which could be induced by binding of the endogenous ligand, cGAMP, to the cytosolic ligand-binding domain. Here we report the discovery through functional screens of a class of compounds, named NVS-STGs, that activate human STING. Our cryo-EM structures show that NVS-STG2 induces the high-order oligomerization of human STING by binding to a pocket between the transmembrane domains of the neighboring STING dimers, effectively acting as a molecular glue. Our functional assays showed that NVS-STG2 could elicit potent STING-mediated immune responses in cells and antitumor activities in animal models. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29281.map.gz emd_29281.map.gz | 4.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29281-v30.xml emd-29281-v30.xml emd-29281.xml emd-29281.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29281_fsc.xml emd_29281_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_29281.png emd_29281.png | 91.3 KB | ||
| Filedesc metadata |  emd-29281.cif.gz emd-29281.cif.gz | 6.6 KB | ||
| Others |  emd_29281_half_map_1.map.gz emd_29281_half_map_1.map.gz emd_29281_half_map_2.map.gz emd_29281_half_map_2.map.gz | 11.1 MB 11.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29281 http://ftp.pdbj.org/pub/emdb/structures/EMD-29281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29281 | HTTPS FTP |
-Related structure data
| Related structure data |  8flkMC  8flmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29281.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29281.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of STING tetramer bound to cGAMP and NVS-STG2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 1
| File | emd_29281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_29281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : STING tetramer
| Entire | Name: STING tetramer |
|---|---|
| Components |
|
-Supramolecule #1: STING tetramer
| Supramolecule | Name: STING tetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: STING tetramer, which can further assemblies into larger oligomers, induced by cGAMP and NVS-STG2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Stimulator of interferon genes protein
| Macromolecule | Name: Stimulator of interferon genes protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.553398 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPHSSLHPSI PCPRGHGAQK AALVLLSACL VTLWGLGEPP EHTLRYLVLH LASLQLGLLL NGVCSLAEEL RHIHSRYRGS YWRTVRACL GCPLRRGALL LLSIYFYYSL PNAVGPPFTW MLALLGLSQA LNILLGLKGL APAEISAVCE KGNFNVAHGL A WSYYIGYL ...String: MPHSSLHPSI PCPRGHGAQK AALVLLSACL VTLWGLGEPP EHTLRYLVLH LASLQLGLLL NGVCSLAEEL RHIHSRYRGS YWRTVRACL GCPLRRGALL LLSIYFYYSL PNAVGPPFTW MLALLGLSQA LNILLGLKGL APAEISAVCE KGNFNVAHGL A WSYYIGYL RLILPELQAR IRTYNQHYNN LLRGAVSQRL YILLPLDCGV PDNLSMADPN IRFLDKLPQQ TGDRAGIKDR VY SNSIYEL LENGQRAGTC VLEYATPLQT LFAMSQYSQA GFSREDRLEQ AKLFCRTLED ILADAPESQN NCRLIAYQEP ADD SSFSLS QEVLRHLRQE EKEEVTVGTS SGLEVLFQ UniProtKB: Stimulator of interferon genes protein |
-Macromolecule #2: cGAMP
| Macromolecule | Name: cGAMP / type: ligand / ID: 2 / Number of copies: 2 / Formula: 1SY |
|---|---|
| Molecular weight | Theoretical: 674.411 Da |
| Chemical component information | 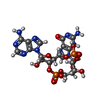 ChemComp-1SY: |
-Macromolecule #3: 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}a...
| Macromolecule | Name: 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid type: ligand / ID: 3 / Number of copies: 2 / Formula: Y6H |
|---|---|
| Molecular weight | Theoretical: 427.533 Da |
| Chemical component information | 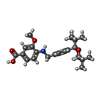 ChemComp-Y6H: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8flk: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)