[English] 日本語
 Yorodumi
Yorodumi- EMDB-10898: Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10898 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit | |||||||||
 Map data Map data | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, post-processed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibiotic / tigecycline / translation / ribosome | |||||||||
| Function / homology |  Function and homology information Function and homology informationassembly of large subunit precursor of preribosome / large ribosomal subunit / transferase activity / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding ...assembly of large subunit precursor of preribosome / large ribosomal subunit / transferase activity / ribosome binding / ribosomal large subunit assembly / 5S rRNA binding / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) / Acinetobacter baumannii (bacteria) /  Acinetobacter baumannii ATCC 19606 = CIP 70.34 = JCM 6841 (bacteria) Acinetobacter baumannii ATCC 19606 = CIP 70.34 = JCM 6841 (bacteria) | |||||||||
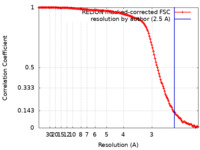
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Nicholson D / Edwards TA | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics. Authors: David Nicholson / Thomas A Edwards / Alex J O'Neill / Neil A Ranson /  Abstract: Acinetobacter baumannii is a Gram-negative bacterium primarily associated with hospital-acquired, often multidrug-resistant (MDR) infections. The ribosome-targeting antibiotics amikacin and ...Acinetobacter baumannii is a Gram-negative bacterium primarily associated with hospital-acquired, often multidrug-resistant (MDR) infections. The ribosome-targeting antibiotics amikacin and tigecycline are among the limited arsenal of drugs available for treatment of such infections. We present high-resolution structures of the 70S ribosome from A. baumannii in complex with these antibiotics, as determined by cryoelectron microscopy. Comparison with the ribosomes of other bacteria reveals several unique structural features at functionally important sites, including around the exit of the polypeptide tunnel and the periphery of the subunit interface. The structures also reveal the mode and site of interaction of these drugs with the ribosome. This work paves the way for the design of new inhibitors of translation to address infections caused by MDR A. baumannii. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10898.map.gz emd_10898.map.gz | 29.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10898-v30.xml emd-10898-v30.xml emd-10898.xml emd-10898.xml | 51.7 KB 51.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10898_fsc.xml emd_10898_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10898.png emd_10898.png | 32.7 KB | ||
| Masks |  emd_10898_msk_1.map emd_10898_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10898.cif.gz emd-10898.cif.gz | 10.4 KB | ||
| Others |  emd_10898_additional.map.gz emd_10898_additional.map.gz emd_10898_half_map_1.map.gz emd_10898_half_map_1.map.gz emd_10898_half_map_2.map.gz emd_10898_half_map_2.map.gz | 141.3 MB 182.8 MB 182.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10898 http://ftp.pdbj.org/pub/emdb/structures/EMD-10898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10898 | HTTPS FTP |
-Related structure data
| Related structure data |  6ysiMC  6yhsC  6ypuC  6ys5C  6yt9C  6ytfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10407 (Title: Motion-corrected micrographs and extracted particle images of the 70S ribosome from the human pathogen Acinetobacter baumannii in complex with tigecycline EMPIAR-10407 (Title: Motion-corrected micrographs and extracted particle images of the 70S ribosome from the human pathogen Acinetobacter baumannii in complex with tigecyclineData size: 538.5 Data #1: Motion-corrected micrographs of the 70S ribosome from the human pathogen Acinetobacter baumannii in complex with tigecycline [micrographs - single frame] Data #2: Extracted particle images of the 70S ribosome from the human pathogen Acinetobacter baumannii in complex with tigecycline [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10898.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10898.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
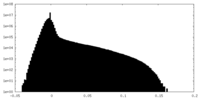
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10898_msk_1.map emd_10898_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
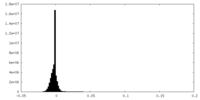
| Density Histograms |
-Additional map: Acinetobacter baumannii ribosome-tigecycline complex, consensus map filtered by...
| File | emd_10898_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acinetobacter baumannii ribosome-tigecycline complex, consensus map filtered by local resolution | ||||||||||||
| Projections & Slices |
| ||||||||||||
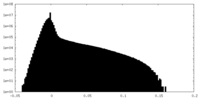
| Density Histograms |
-Half map: Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, half...
| File | emd_10898_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, half...
| File | emd_10898_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit
+Supramolecule #1: Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit
+Macromolecule #1: 50S ribosomal protein L2
+Macromolecule #2: 50S ribosomal protein L3
+Macromolecule #3: 50S ribosomal protein L4
+Macromolecule #4: 50S ribosomal protein L5
+Macromolecule #5: 50S ribosomal protein L6
+Macromolecule #6: 50S ribosomal protein L13
+Macromolecule #7: 50S ribosomal protein L14
+Macromolecule #8: 50S ribosomal protein L15
+Macromolecule #9: 50S ribosomal protein L16
+Macromolecule #10: 50S ribosomal protein L17
+Macromolecule #11: 50S ribosomal protein L18
+Macromolecule #12: 50S ribosomal protein L19
+Macromolecule #13: 50S ribosomal protein L20
+Macromolecule #14: 50S ribosomal protein L21
+Macromolecule #15: 50S ribosomal protein L22
+Macromolecule #16: 50S ribosomal protein L23
+Macromolecule #17: 50S ribosomal protein L24
+Macromolecule #18: 50S ribosomal protein L25
+Macromolecule #19: 50S ribosomal protein L27
+Macromolecule #20: 50S ribosomal protein L28
+Macromolecule #21: 50S ribosomal protein L29
+Macromolecule #22: 50S ribosomal protein L30
+Macromolecule #23: 50S ribosomal protein L31
+Macromolecule #24: 50S ribosomal protein L32
+Macromolecule #25: 50S ribosomal protein L33
+Macromolecule #26: 50S ribosomal protein L34
+Macromolecule #30: 50S ribosomal protein L35
+Macromolecule #31: 50S ribosomal protein L36
+Macromolecule #27: 23S ribosomal RNA
+Macromolecule #28: 5S ribosomal RNA
+Macromolecule #29: E-site tRNA
+Macromolecule #32: MAGNESIUM ION
+Macromolecule #33: TIGECYCLINE
+Macromolecule #34: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 1.1 sec. / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient | ||||||
| Output model |  PDB-6ysi: |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)