+Search query
-Structure paper
| Title | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics. |
|---|---|
| Journal, issue, pages | Structure, Vol. 28, Issue 10, Page 1087-11100.e3, Year 2020 |
| Publish date | Oct 6, 2020 |
 Authors Authors | David Nicholson / Thomas A Edwards / Alex J O'Neill / Neil A Ranson /  |
| PubMed Abstract | Acinetobacter baumannii is a Gram-negative bacterium primarily associated with hospital-acquired, often multidrug-resistant (MDR) infections. The ribosome-targeting antibiotics amikacin and ...Acinetobacter baumannii is a Gram-negative bacterium primarily associated with hospital-acquired, often multidrug-resistant (MDR) infections. The ribosome-targeting antibiotics amikacin and tigecycline are among the limited arsenal of drugs available for treatment of such infections. We present high-resolution structures of the 70S ribosome from A. baumannii in complex with these antibiotics, as determined by cryoelectron microscopy. Comparison with the ribosomes of other bacteria reveals several unique structural features at functionally important sites, including around the exit of the polypeptide tunnel and the periphery of the subunit interface. The structures also reveal the mode and site of interaction of these drugs with the ribosome. This work paves the way for the design of new inhibitors of translation to address infections caused by MDR A. baumannii. |
 External links External links |  Structure / Structure /  PubMed:32857965 / PubMed:32857965 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.5 - 3.0 Å |
| Structure data | EMDB-10809, PDB-6yhs: EMDB-10869, PDB-6ypu: EMDB-10892, PDB-6ys5: EMDB-10898, PDB-6ysi: EMDB-10914, PDB-6yt9: EMDB-10915, PDB-6ytf: |
| Chemicals |  ChemComp-MG:  ChemComp-ZN: 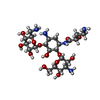 ChemComp-AKN: 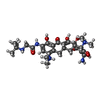 ChemComp-T1C: |
| Source |
|
 Keywords Keywords | RIBOSOME / antibiotic / amikacin / translation / tigecycline |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841 (bacteria)
acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841 (bacteria)