-Search query
-Search result
Showing 1 - 50 of 56 items for (author: mehta & v)

EMDB-16255: 
Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions

PDB-8bv5: 
Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions

EMDB-16249: 
Structure of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc

EMDB-16252: 
Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-16253: 
Focused refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-16254: 
Focused refinement of transmembrane domain of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

PDB-8buz: 
Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-17182: 
Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in GDN

EMDB-17185: 
Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP

EMDB-17186: 
Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in nanodisc

PDB-8otq: 
Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in GDN

PDB-8otw: 
Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP

PDB-8otx: 
Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in nanodisc

EMDB-34813: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril

PDB-8hia: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril

EMDB-16603: 
Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16604: 
Alpha-synuclein filament assembled in vitro with mutant (7 residues insertion) protein

EMDB-16608: 
WT alpha-synuclein filament assembled in vitro

PDB-8ceb: 
Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16600: 
Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

PDB-8ce7: 
Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16188: 
Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy

EMDB-16189: 
Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy

PDB-8bqv: 
Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy

PDB-8bqw: 
Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy

EMDB-14388: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya

PDB-7yzi: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya

EMDB-14389: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya

PDB-7yzk: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya

EMDB-14174: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)

EMDB-14176: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)

EMDB-14187: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)

EMDB-14188: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)

EMDB-14189: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)
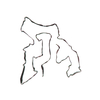
PDB-7qvc: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)
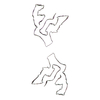
PDB-7qvf: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)
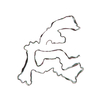
PDB-7qwg: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)
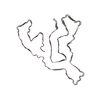
PDB-7qwl: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)

PDB-7qwm: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)

EMDB-13336: 
focus refinement of soluble domain of adenylyl cyclase 9 in complex with Gs protein alpha subunit and MANT-GTP

PDB-7pdf: 
focus refinement of soluble domain of adenylyl cyclase 9 in complex with Gs protein alpha subunit and MANT-GTP

EMDB-13330: 
structure of Adenylyl cyclase 9 in complex with MANT-GTP

EMDB-13331: 
Structure of Adenylyl cyclase 9 in complex with DARPin C4 and MANT-GTP

EMDB-13334: 
Focus refinement of soluble domain of Adenylyl cyclase 9 in complex with DARPin C4 and MANT-GTP

EMDB-13335: 
Structure of Adenylyl cyclase 9 in complex with Gs protein alpha subunit and MANT-GTP

EMDB-13337: 
structure of adenylyl cyclase 9 in complex with DARPin C4 and ATP-aS

EMDB-13338: 
structure of adenylyl cyclase 9 in complex with DARPin C4 and ATP-aS

PDB-7pd4: 
structure of Adenylyl cyclase 9 in complex with MANT-GTP

PDB-7pd8: 
Structure of Adenylyl cyclase 9 in complex with DARPin C4 and MANT-GTP

PDB-7pdd: 
Focus refinement of soluble domain of Adenylyl cyclase 9 in complex with DARPin C4 and MANT-GTP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
