-Search query
-Search result
Showing all 35 items for (author: lucic & v)

EMDB-52343: 
Native intermediate synaptic vesicle tether, likely Munc13
Method: subtomogram averaging / : Lucic V, Oroczo-Borunda DH

EMDB-52342: 
Short (SNARE complex dependent) synaptic vesicle tether
Method: subtomogram averaging / : Lucic V, Oroczo-Borunda DH

EMDB-18728: 
Rat synaptosome
Method: electron tomography / : Radecke J, Seeger R, Zuber B

EMDB-18744: 
Stimulated rat synaptosome.
Method: electron tomography / : Radecke J, Seeger R, Zuber B

EMDB-18746: 
wild type neuronal synapse
Method: electron tomography / : Radecke J, Seeger R, Zuber B, Sorensen JB

EMDB-18748: 
SNAP-25-4E neuronal synapse
Method: electron tomography / : Radecke J, Seeger R, Zuber B, Sorensen JB

EMDB-18749: 
SNAP-25-4K neuronal synapse
Method: electron tomography / : Radecke J, Seeger R, Zuber B, Sorensen JB

EMDB-16083: 
Munc13-SNAP25 cryo-ET dataset, synapse tomo WT 108 (id: m13_ctrl_108)
Method: electron tomography / : Papantoniou C, Lucic V

EMDB-16084: 
Munc13-SNAP25 cryo-ET dataset, synapse tomo Munc13 DHet 115 (id: m13_dhet_115)
Method: electron tomography / : Papantoniou C, Lucic V

EMDB-16085: 
Munc13-SNAP25 cryo-ET dataset, synapse tomo Munc13 DKO 102 (id: m13_dko_102)
Method: electron tomography / : Papantoniou C, Lucic V

EMDB-11404: 
Native putative AMPA-type ionotropic glutamate receptor, de novo
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-11405: 
Native putative NMDA-type ionotropic glutamate receptor, de novo
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-11406: 
Native putative AMPAR-type glutamate receptor, de novo 3D classification
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-11407: 
Native putative NMDAR type glutaamate receptor, de novo 3D classification
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-11408: 
Native putative AMPAR-type glutamate receptor, de novo 3D classification C2
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-11409: 
Native putative NMDAR-type glutamate receptor, de novo 3D classification C2
Method: electron tomography / : Martinez A, Lucic V

EMDB-11410: 
Native putative AMPAR type glutamate receptor, model guided
Method: electron tomography / : Martinez A, Lucic V

EMDB-11411: 
Native putative NMDAR type glutamate receptor, model guided
Method: subtomogram averaging / : Martinez A, Lucic V

EMDB-10432: 
In situ ER membrane bound ribosome
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10433: 
In situ ER membrane bound translocon-ribosome
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10434: 
In situ ER membrane bound ribosome-free translocon
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10435: 
Putative in situ ER membrane bound major histocompatibility peptide loading complex (PLC)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S
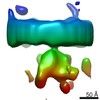
EMDB-10436: 
Putative in situ ER membrane bound major histocompatibility peptide loading complex (PLC)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S
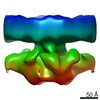
EMDB-10437: 
Putative in situ ER membrane bound inositol trisphosphate receptor complex (IP3R)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10439: 
Example of tomogram used for in situ template-free membrane bound complexes determination
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10449: 
ER Microsome in Fig. 3 (Martinez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-10450: 
Endoplasmic Reticulum microsome used for Videos in (Martinez-Sanchez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-10451: 
ER microsome released with the code in (Martinez-Sanchez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-0074: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0075: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0084: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0085: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0086: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0087: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
