[English] 日本語
 Yorodumi
Yorodumi- EMDB-1394: Snapshots of nuclear pore complexes in action captured by cryo-el... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1394 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
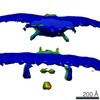
| Title | Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. | |||||||||
 Map data Map data | asymmetric unit of Dictyostelium discoideum nuclear pore complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 58.0 Å | |||||||||
 Authors Authors | Beck M / Lucic V / Forster F / Baumeister W / Medalia O | |||||||||
 Citation Citation |  Journal: Nature / Year: 2007 Journal: Nature / Year: 2007Title: Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Authors: Martin Beck / Vladan Lucić / Friedrich Förster / Wolfgang Baumeister / Ohad Medalia /  Abstract: Nuclear pore complexes reside in the nuclear envelope of eukaryotic cells and mediate the nucleocytoplasmic exchange of macromolecules. Traffic is regulated by mobile transport receptors that target ...Nuclear pore complexes reside in the nuclear envelope of eukaryotic cells and mediate the nucleocytoplasmic exchange of macromolecules. Traffic is regulated by mobile transport receptors that target their cargo to the central translocation channel, where phenylalanine-glycine-rich repeats serve as binding sites. The structural analysis of the nuclear pore is a formidable challenge given its size, its location in a membranous environment and its dynamic nature. Here we have used cryo-electron tomography to study the structure of nuclear pore complexes in their functional environment, that is, in intact nuclei of Dictyostelium discoideum. A new image-processing strategy compensating for deviations of the asymmetric units (protomers) from a perfect eight-fold symmetry enabled us to refine the structure and to identify new features. Furthermore, the superposition of a large number of tomograms taken in the presence of cargo, which was rendered visible by gold nanoparticles, has yielded a map outlining the trajectories of import cargo. Finally, we have performed single-molecule Monte Carlo simulations of nuclear import to interpret the experimentally observed cargo distribution in the light of existing models for nuclear import. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1394.map.gz emd_1394.map.gz | 684.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1394-v30.xml emd-1394-v30.xml emd-1394.xml emd-1394.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1394.png EMD-1394.png | 273.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1394 http://ftp.pdbj.org/pub/emdb/structures/EMD-1394 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1394 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1394 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1394.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1394.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | asymmetric unit of Dictyostelium discoideum nuclear pore complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 16.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : nuclear pore complex
| Entire | Name: nuclear pore complex |
|---|---|
| Components |
|
-Supramolecule #1000: nuclear pore complex
| Supramolecule | Name: nuclear pore complex / type: sample / ID: 1000 Oligomeric state: octamer (actual refined structure represents an asymmetric unit of the NPC) Number unique components: 1 |
|---|
-Supramolecule #1: nuclear pore
| Supramolecule | Name: nuclear pore / type: organelle_or_cellular_component / ID: 1 / Name.synonym: nuclear pore / Oligomeric state: octamer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Details: 50 mM Tris pH 7.6, 25 mM KCl, 5mM MgCl2, 50 mM sucrose |
|---|---|
| Grid | Details: 200 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Rudolf Gatz plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 90 K |
| Specialist optics | Energy filter - Name: GIF2002 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -63 ° / Tilt series - Axis1 - Max angle: 63 ° |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Average number of projections used in the 3D reconstructions: 4184. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 58.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: TOM |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















