[English] 日本語
 Yorodumi
Yorodumi- EMDB-0085: Template-free detection and classification of microsomal membrane... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Template-free detection and classification of microsomal membrane bound complexes | |||||||||
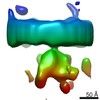
 Map data Map data | Translocon complex associated with ribosome | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 22.0 Å | |||||||||
 Authors Authors | Martinez-Sanchez A / Lucic V | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Structure of the native Sec61 protein-conducting channel. Authors: Stefan Pfeffer / Laura Burbaum / Pia Unverdorben / Markus Pech / Yuxiang Chen / Richard Zimmermann / Roland Beckmann / Friedrich Förster /  Abstract: In mammalian cells, secretory and membrane proteins are translocated across or inserted into the endoplasmic reticulum (ER) membrane by the universally conserved protein-conducting channel Sec61, ...In mammalian cells, secretory and membrane proteins are translocated across or inserted into the endoplasmic reticulum (ER) membrane by the universally conserved protein-conducting channel Sec61, which has been structurally studied in isolated, detergent-solubilized states. Here we structurally and functionally characterize native, non-solubilized ribosome-Sec61 complexes on rough ER vesicles using cryo-electron tomography and ribosome profiling. Surprisingly, the 9-Å resolution subtomogram average reveals Sec61 in a laterally open conformation, even though the channel is not in the process of inserting membrane proteins into the lipid bilayer. In contrast to recent mechanistic models for polypeptide translocation and insertion, our results indicate that the laterally open conformation of Sec61 is the only conformation present in the ribosome-bound translocon complex, independent of its functional state. Consistent with earlier functional studies, our structure suggests that the ribosome alone, even without a nascent chain, is sufficient for lateral opening of Sec61 in a lipid environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0085.map.gz emd_0085.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0085-v30.xml emd-0085-v30.xml emd-0085.xml emd-0085.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0085_fsc.xml emd_0085_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0085.png emd_0085.png | 38.1 KB | ||
| Others |  emd_0085_half_map_1.map.gz emd_0085_half_map_1.map.gz emd_0085_half_map_2.map.gz emd_0085_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0085 http://ftp.pdbj.org/pub/emdb/structures/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0085 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0085.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0085.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Translocon complex associated with ribosome | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||
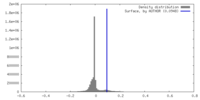
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Translocon complex associated with ribosome, 1st half
| File | emd_0085_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Translocon complex associated with ribosome, 1st half | ||||||||||||
| Projections & Slices |
| ||||||||||||
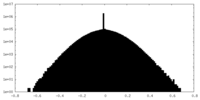
| Density Histograms |
-Half map: Translocon complex associated with ribosome, 2nd half
| File | emd_0085_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Translocon complex associated with ribosome, 2nd half | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mammalian ribosome bound to the native protein translocon on cani...
| Entire | Name: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles |
|---|---|
| Components |
|
-Supramolecule #1: Mammalian ribosome bound to the native protein translocon on cani...
| Supramolecule | Name: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles type: complex / ID: 1 / Parent: 0 / Details: The same sample as the one used for EMD-3068 - 72 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: The same as for EMD-3068 - 72 | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 70 % / Instrument: FEI VITROBOT MARK IV / Details: Blot 3 seconds before plunging.. | ||||||||||||
| Details | The same sample was used for EMD-3068 - 72 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | The same as for EMD-3068 - 72 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 0.75 e/Å2 / Details: The same as for EMD-3068 - 72 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller












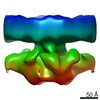










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)