+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9907 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
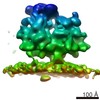
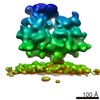
| Title | RdRp complex | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cypovirus / Transcription / RNA-dependent RNA polymerase / VIRAL PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=2 icosahedral viral capsid / viral inner capsid / viral genome replication / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Cypovirus 1 / Cypovirus 1 /  Cypovirus (cytoplasmic polyhedrosis viruses) Cypovirus (cytoplasmic polyhedrosis viruses) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Li XW | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Structure of RdRps Within a Transcribing dsRNA Virus Provides Insights Into the Mechanisms of RNA Synthesis. Authors: Xiaowu Li / Li Wang / Xurong Wang / Wenyuan Chen / Tao Yang / Jingdong Song / Hongrong Liu / Lingpeng Cheng /  Abstract: RNA-dependent RNA polymerases (RdRps) catalyze RNA synthesis of RNA viruses. During initiation of RNA synthesis, the RdRp catalyzes the formation of the first dinucleotide, acting as primer for ...RNA-dependent RNA polymerases (RdRps) catalyze RNA synthesis of RNA viruses. During initiation of RNA synthesis, the RdRp catalyzes the formation of the first dinucleotide, acting as primer for subsequent processive RNA elongation. Here, we present the structure of the RdRp complexes in the dinucleotide primed state in situ within a transcribing cypovirus under near physiological conditions using cryo-electron microscopy. The 3' end of RNA templates, paired RNA dinucleotide primer, incoming nucleotide, and catalytic divalent cations in the RdRp were resolved at 3.8 Å resolution. The end of the RNA template and the dinucleotide is buttressed by the aromatic tyrosine in a loop from the RdRp bracelet domain. Our structure reveals the interactions between the nucleotide substrates and the conserved residues during the RdRp initiation, and the coordinated structural changes preceding the elongation stage. In addition, it provides the direct evidence for existence of the slow step of the dinucleotide primed state in the viral RdRp transcription. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9907.map.gz emd_9907.map.gz | 40.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9907-v30.xml emd-9907-v30.xml emd-9907.xml emd-9907.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9907.png emd_9907.png | 135.1 KB | ||
| Filedesc metadata |  emd-9907.cif.gz emd-9907.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9907 http://ftp.pdbj.org/pub/emdb/structures/EMD-9907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9907 | HTTPS FTP |
-Related structure data
| Related structure data |  6k32MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9907.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9907.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
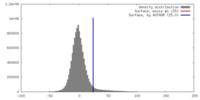
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cypovirus
+Supramolecule #1: Cypovirus
+Macromolecule #1: RNA-dependent RNA polymerase
+Macromolecule #2: Viral structural protein 4
+Macromolecule #5: VP1
+Macromolecule #6: VP1
+Macromolecule #7: VP1
+Macromolecule #3: RNA (5'-R(P*UP*UP*AP*CP*U)-3')
+Macromolecule #4: RNA (5'-D(*(3PO))-R(*AP*G)-3'
+Macromolecule #8: MAGNESIUM ION
+Macromolecule #9: 7-METHYLGUANOSINE
+Macromolecule #10: DIPHOSPHATE
+Macromolecule #11: 2'-O-methyladenosine 5'-(dihydrogen phosphate)
+Macromolecule #12: URIDINE 5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



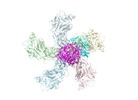





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















 invertebrate environmental sample (environmental samples)
invertebrate environmental sample (environmental samples)