[English] 日本語
 Yorodumi
Yorodumi- EMDB-0074: Template-free detection and classification of microsomal membrane... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Template-free detection and classification of microsomal membrane bound complexes | |||||||||
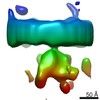
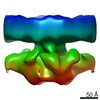
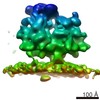
 Map data Map data | Ribosome bound to fully assembled translocon complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
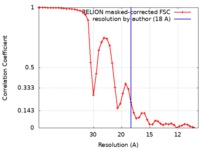
| Method | subtomogram averaging / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Martinez-Sanchez A / Lucic V | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2020 Journal: Nat Methods / Year: 2020Title: Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms. Authors: Antonio Martinez-Sanchez / Zdravko Kochovski / Ulrike Laugks / Johannes Meyer Zum Alten Borgloh / Saikat Chakraborty / Stefan Pfeffer / Wolfgang Baumeister / Vladan Lučić /  Abstract: With faithful sample preservation and direct imaging of fully hydrated biological material, cryo-electron tomography provides an accurate representation of molecular architecture of cells. However, ...With faithful sample preservation and direct imaging of fully hydrated biological material, cryo-electron tomography provides an accurate representation of molecular architecture of cells. However, detection and precise localization of macromolecular complexes within cellular environments is aggravated by the presence of many molecular species and molecular crowding. We developed a template-free image processing procedure for accurate tracing of complex networks of densities in cryo-electron tomograms, a comprehensive and automated detection of heterogeneous membrane-bound complexes and an unsupervised classification (PySeg). Applications to intact cells and isolated endoplasmic reticulum (ER) allowed us to detect and classify small protein complexes. This classification provided sufficiently homogeneous particle sets and initial references to allow subsequent de novo subtomogram averaging. Spatial distribution analysis showed that ER complexes have different localization patterns forming nanodomains. Therefore, this procedure allows a comprehensive detection and structural analysis of complexes in situ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0074.map.gz emd_0074.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0074-v30.xml emd-0074-v30.xml emd-0074.xml emd-0074.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0074_fsc.xml emd_0074_fsc.xml | 4.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_0074.png emd_0074.png | 36.7 KB | ||
| Others |  emd_0074_half_map_1.map.gz emd_0074_half_map_1.map.gz emd_0074_half_map_2.map.gz emd_0074_half_map_2.map.gz | 4.9 MB 5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0074 http://ftp.pdbj.org/pub/emdb/structures/EMD-0074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0074 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0074.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0074.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ribosome bound to fully assembled translocon complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Ribosome bound to fully assembled translocon complex, 2nd half map
| File | emd_0074_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ribosome bound to fully assembled translocon complex, 2nd half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Ribosome bound to fully assembled translocon complex, 1st half map
| File | emd_0074_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ribosome bound to fully assembled translocon complex, 1st half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mammalian ribosome bound to the native protein translocon on cani...
| Entire | Name: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles |
|---|---|
| Components |
|
-Supramolecule #1: Mammalian ribosome bound to the native protein translocon on cani...
| Supramolecule | Name: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 70 % / Instrument: FEI VITROBOT MARK IV / Details: Blot 3 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)