-Search query
-Search result
Showing all 44 items for (author: liu & yz)

EMDB-36869: 
Cryo-EM structure of CXCR4 in complex with CXCL12

EMDB-38596: 
Cryo-EM structure of cryptophyte photosystem II

EMDB-37416: 
Cryo-EM structure of Snf7 N-terminal domain in outer coils of spiral polymers

EMDB-37417: 
CryoEM structure of Snf7 N-terminal domain in the inner coils of spiral
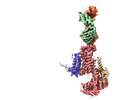
EMDB-37159: 
Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS

EMDB-36366: 
Cryo-EM structure of Symbiodinium photosystem I
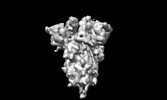
EMDB-35239: 
Prefusion spike of BA.1 with all-RBD-down conformation

EMDB-33209: 
AhCS-3Mg2+-FSPP

EMDB-34338: 
Postfusion spike of BA.1.1

EMDB-34339: 
Prefusion spike protein of BA.1.1 with one-RBD-up conformation

EMDB-34340: 
Prefusion spike of BA.2.3 with one-RBD-up conformation

EMDB-34341: 
Postfusion spike of BA.2.3

EMDB-34343: 
Prefusion spike of BA.2.3 with all-RBD-down conformation

EMDB-33422: 
structure of a membrane-integrated glycosyltransferase with inhibitor

EMDB-33423: 
structure of a membrane-integrated glycosyltransferase

EMDB-33404: 
3.4 Angstrom cryoEM D5 reconstruction of mud crab reovirus

EMDB-33659: 
Cryo-EM structure of cryptophyte photosystem I

EMDB-33683: 
Cryo-EM structure of cryptophyte photosystem I

EMDB-33142: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab

EMDB-32356: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine

EMDB-33498: 
Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A

EMDB-33153: 
Cryo-EM structure of plant NLR Sr35 resistosome

EMDB-33486: 
Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35

EMDB-11953: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map

EMDB-11954: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)

EMDB-14810: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map

EMDB-14811: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement

EMDB-31824: 
Cryo-EM structure of the SV1-Gs complex.

EMDB-32908: 
Flagellar motor from wild type H. pylori

EMDB-32909: 
Flagellar motor of H.pylori delta-FliY mutant

EMDB-32910: 
Flagellar motor of H.pylori FliYc mutant

EMDB-25143: 
Structure of positive allosteric modulator-bound active human calcium-sensing receptor

EMDB-25144: 
Structure of positive allosteric modulator-free active human calcium-sensing receptor

EMDB-25145: 
Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor

EMDB-20840: 
FACT_subnucleosome complex 1

EMDB-20841: 
FACT_subnucleosome complex 2

EMDB-7092: 
Cryo-EM structure of human insulin degrading enzyme in complex with insulin

EMDB-7090: 
Cryo-EM structure of human insulin degrading enzyme

EMDB-7091: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain

EMDB-7093: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain

EMDB-7066: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain

EMDB-7062: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin

EMDB-7041: 
Cryo-EM structure of human insulin degrading enzyme in complex with insulin

EMDB-7065: 
Cryo-EM structure of human insulin degrading enzyme in complex with insulin
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
