[English] 日本語
 Yorodumi
Yorodumi- EMDB-25143: Structure of positive allosteric modulator-bound active human cal... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25143 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | calcium-sensing receptor / cryo-EM structure / allosteric modulation / activation mechanism / symmetry / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology | Isoform 1 of Extracellular calcium-sensing receptor Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||
 Authors Authors | Park J / Zuo H | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Symmetric activation and modulation of the human calcium-sensing receptor. Authors: Jinseo Park / Hao Zuo / Aurel Frangaj / Ziao Fu / Laura Y Yen / Zhening Zhang / Lidia Mosyak / Vesna N Slavkovich / Jonathan Liu / Kimberly M Ray / Baohua Cao / Francesca Vallese / Yong Geng ...Authors: Jinseo Park / Hao Zuo / Aurel Frangaj / Ziao Fu / Laura Y Yen / Zhening Zhang / Lidia Mosyak / Vesna N Slavkovich / Jonathan Liu / Kimberly M Ray / Baohua Cao / Francesca Vallese / Yong Geng / Shaoxia Chen / Robert Grassucci / Venkata P Dandey / Yong Zi Tan / Edward Eng / Yeji Lee / Brian Kloss / Zheng Liu / Wayne A Hendrickson / Clinton S Potter / Bridget Carragher / Joseph Graziano / Arthur D Conigrave / Joachim Frank / Oliver B Clarke / Qing R Fan /     Abstract: The human extracellular calcium-sensing (CaS) receptor controls plasma Ca levels and contributes to nutrient-dependent maintenance and metabolism of diverse organs. Allosteric modulation of the CaS ...The human extracellular calcium-sensing (CaS) receptor controls plasma Ca levels and contributes to nutrient-dependent maintenance and metabolism of diverse organs. Allosteric modulation of the CaS receptor corrects disorders of calcium homeostasis. Here, we report the cryogenic-electron microscopy reconstructions of a near-full-length CaS receptor in the absence and presence of allosteric modulators. Activation of the homodimeric CaS receptor requires a break in the transmembrane 6 (TM6) helix of each subunit, which facilitates the formation of a TM6-mediated homodimer interface and expansion of homodimer interactions. This transformation in TM6 occurs without a positive allosteric modulator. Two modulators with opposite functional roles bind to overlapping sites within the transmembrane domain through common interactions, acting to stabilize distinct rotamer conformations of key residues on the TM6 helix. The positive modulator reinforces TM6 distortion and maximizes subunit contact to enhance receptor activity, while the negative modulator strengthens an intact TM6 to dampen receptor function. In both active and inactive states, the receptor displays symmetrical transmembrane conformations that are consistent with its homodimeric assembly. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25143.map.gz emd_25143.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25143-v30.xml emd-25143-v30.xml emd-25143.xml emd-25143.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25143.png emd_25143.png | 117.1 KB | ||
| Filedesc metadata |  emd-25143.cif.gz emd-25143.cif.gz | 7.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25143 http://ftp.pdbj.org/pub/emdb/structures/EMD-25143 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25143 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25143 | HTTPS FTP |
-Validation report
| Summary document |  emd_25143_validation.pdf.gz emd_25143_validation.pdf.gz | 408.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25143_full_validation.pdf.gz emd_25143_full_validation.pdf.gz | 408 KB | Display | |
| Data in XML |  emd_25143_validation.xml.gz emd_25143_validation.xml.gz | 4.2 KB | Display | |
| Data in CIF |  emd_25143_validation.cif.gz emd_25143_validation.cif.gz | 4.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25143 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25143 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25143 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25143 | HTTPS FTP |
-Related structure data
| Related structure data |  7silMC  7simC  7sinC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10834 (Title: Structures of positive allosteric modulator-bound and unbound active human calcium-sensing receptor EMPIAR-10834 (Title: Structures of positive allosteric modulator-bound and unbound active human calcium-sensing receptorData size: 3.8 TB Data #1: Unaligned multi-frame micrographs of calcium-sensing receptor in both PAM-bound and unbound active states [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25143.map.gz / Format: CCP4 / Size: 16 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25143.map.gz / Format: CCP4 / Size: 16 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Homodimer human calcium sensing receptor
| Entire | Name: Homodimer human calcium sensing receptor |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer human calcium sensing receptor
| Supramolecule | Name: Homodimer human calcium sensing receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 192 KDa |
-Macromolecule #1: Isoform 1 of Extracellular calcium-sensing receptor
| Macromolecule | Name: Isoform 1 of Extracellular calcium-sensing receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.299461 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFYSCCWVL LALTWHTSAY GPDQRAQKKG DIILGGLFPI HFGVAAKDQD LKSRPESVEC IRYNFRGFRW LQAMIFAIEE INSSPALLP NLTLGYRIFD TCNTVSKALE ATLSFVAQNK IDSLNLDEFC NCSEHIPSTI AVVGATGSGV STAVANLLGL F YIPQVSYA ...String: MAFYSCCWVL LALTWHTSAY GPDQRAQKKG DIILGGLFPI HFGVAAKDQD LKSRPESVEC IRYNFRGFRW LQAMIFAIEE INSSPALLP NLTLGYRIFD TCNTVSKALE ATLSFVAQNK IDSLNLDEFC NCSEHIPSTI AVVGATGSGV STAVANLLGL F YIPQVSYA SSSRLLSNKN QFKSFLRTIP NDEHQATAMA DIIEYFRWNW VGTIAADDDY GRPGIEKFRE EAEERDICID FS ELISQYS DEEEIQHVVE VIQNSTAKVI VVFSSGPDLE PLIKEIVRRN ITGKIWLASE AWASSSLIAM PQYFHVVGGT IGF ALKAGQ IPGFREFLKK VHPRKSVHNG FAKEFWEETF NCHLQEGAKG PLPVDTFLRG HEESGDRFSN SSTAFRPLCT GDEN ISSVE TPYIDYTHLR ISYNVYLAVY SIAHALQDIY TCLPGRGLFT NGSCADIKKV EAWQVLKHLR HLNFTNNMGE QVTFD ECGD LVGNYSIINW HLSPEDGSIV FKEVGYYNVY AKKGERLFIN EEKILWSGFS REVPFSNCSR DCLAGTRKGI IEGEPT CCF ECVECPDGEY SDETDASACN KCPDDFWSNE NHTSCIAKEI EFLSWTEPFG IALTLFAVLG IFLTAFVLGV FIKFRNT PI VKATNRELSY LLLFSLLCCF SSSLFFIGEP QDWTCRLRQP AFGISFVLCI SCILVKTNRV LLVFEAKIPT SFHRKWWG L NLQFLLVFLC TFMQIVICVI WLYTAPPSSY RNQELEDEII FITCHEGSLM ALGFLIGYTC LLAAICFFFA FKSRKLPEN FNEAKFITFS MLIFFIVWIS FIPAYASTYG KFVSAVEVIA ILAASFGLLA CIFFNKIYII LFKPSRNTIE DYKDDDDK UniProtKB: Isoform 1 of Extracellular calcium-sensing receptor |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: CYCLOMETHYLTRYPTOPHAN
| Macromolecule | Name: CYCLOMETHYLTRYPTOPHAN / type: ligand / ID: 4 / Number of copies: 2 / Formula: TCR |
|---|---|
| Molecular weight | Theoretical: 216.236 Da |
| Chemical component information | 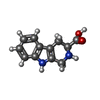 ChemComp-TCR: |
-Macromolecule #5: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine
| Macromolecule | Name: 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine type: ligand / ID: 7 / Number of copies: 2 / Formula: 9IG |
|---|---|
| Molecular weight | Theoretical: 303.826 Da |
| Chemical component information |  ChemComp-9IG: |
-Macromolecule #8: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 8 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solution were made fresh from concentrated, and filtered to avoid microbial contamination. | ||||||||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. | ||||||||||||||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Max: 100.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum ER / Energy filter - Slit width: 20 eV |
| Details | Preliminary grid screening was performed manually. |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 13246 / Average electron dose: 70.35 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-7sil: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















