[English] 日本語
 Yorodumi
Yorodumi- EMDB-31536: Thermostabilised full length human mGluR5-5M bound with L-quisqua... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31536 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thermostabilised full length human mGluR5-5M bound with L-quisqualic acid | ||||||||||||||||||
 Map data Map data | The primary map is the map from Relion after post processing with a shape mask and sharpened with a B-factor of -61 A2 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | G protein coupled receptors / Signal transduction / Metabotropic glutamate receptor 5 (GRM5) / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / operant conditioning / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia ...sensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / operant conditioning / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia / positive regulation of long-term neuronal synaptic plasticity / desensitization of G protein-coupled receptor signaling pathway / positive regulation of sensory perception of pain / negative regulation of excitatory postsynaptic potential / positive regulation of dopamine secretion / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / positive regulation of neural precursor cell proliferation / glutamate receptor activity / nuclear inner membrane / Neurexins and neuroligins / astrocyte projection / response to corticosterone / response to morphine / temperature homeostasis / protein tyrosine kinase activator activity / conditioned place preference / regulation of synaptic transmission, glutamatergic / regulation of long-term synaptic depression / positive regulation of calcium-mediated signaling / protein tyrosine kinase binding / dendritic shaft / response to amphetamine / locomotory behavior / synapse organization / postsynaptic density membrane / G protein-coupled receptor activity / cognition / Schaffer collateral - CA1 synapse / cellular response to amyloid-beta / positive regulation of cytosolic calcium ion concentration / G alpha (q) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / learning or memory / positive regulation of MAPK cascade / response to antibiotic / neuronal cell body / dendrite / regulation of DNA-templated transcription / glutamatergic synapse / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
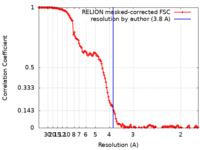
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Vinothkumar KR / Cannone G | ||||||||||||||||||
| Funding support |  India, India,  United Kingdom, United Kingdom,  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations. Authors: Chady Nasrallah / Giuseppe Cannone / Julie Briot / Karine Rottier / Alice E Berizzi / Chia-Ying Huang / Robert B Quast / Francois Hoh / Jean-Louis Banères / Fanny Malhaire / Ludovic Berto / ...Authors: Chady Nasrallah / Giuseppe Cannone / Julie Briot / Karine Rottier / Alice E Berizzi / Chia-Ying Huang / Robert B Quast / Francois Hoh / Jean-Louis Banères / Fanny Malhaire / Ludovic Berto / Anaëlle Dumazer / Joan Font-Ingles / Xavier Gómez-Santacana / Juanlo Catena / Julie Kniazeff / Cyril Goudet / Amadeu Llebaria / Jean-Philippe Pin / Kutti R Vinothkumar / Guillaume Lebon /      Abstract: Metabotropic glutamate receptors (mGluRs) are dimeric G-protein-coupled receptors activated by the main excitatory neurotransmitter, L-glutamate. mGluR activation by agonists binding in the venus ...Metabotropic glutamate receptors (mGluRs) are dimeric G-protein-coupled receptors activated by the main excitatory neurotransmitter, L-glutamate. mGluR activation by agonists binding in the venus flytrap domain is regulated by positive (PAM) or negative (NAM) allosteric modulators binding to the 7-transmembrane domain (7TM). We report the cryo-electron microscopy structures of fully inactive and intermediate-active conformations of mGlu receptor bound to an antagonist and a NAM or an agonist and a PAM, respectively, as well as the crystal structure of the 7TM bound to a photoswitchable NAM. The agonist induces a large movement between the subunits, bringing the 7TMs together and stabilizing a 7TM conformation structurally similar to the inactive state. Using functional approaches, we demonstrate that the PAM stabilizes a 7TM active conformation independent of the conformational changes induced by agonists, representing an alternative mode of mGlu activation. These findings provide a structural basis for different mGluR activation modes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31536.map.gz emd_31536.map.gz | 201.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31536-v30.xml emd-31536-v30.xml emd-31536.xml emd-31536.xml | 28.4 KB 28.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31536_fsc.xml emd_31536_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_31536.png emd_31536.png | 27.9 KB | ||
| Filedesc metadata |  emd-31536.cif.gz emd-31536.cif.gz | 7.9 KB | ||
| Others |  emd_31536_additional_1.map.gz emd_31536_additional_1.map.gz emd_31536_additional_2.map.gz emd_31536_additional_2.map.gz emd_31536_half_map_1.map.gz emd_31536_half_map_1.map.gz emd_31536_half_map_2.map.gz emd_31536_half_map_2.map.gz | 201.7 MB 200.6 MB 169.6 MB 169.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31536 http://ftp.pdbj.org/pub/emdb/structures/EMD-31536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31536 | HTTPS FTP |
-Related structure data
| Related structure data |  7fd8MC  7fd9C  7p2lC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31536.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31536.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The primary map is the map from Relion after post processing with a shape mask and sharpened with a B-factor of -61 A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Postprocessed map with a B-factor of -122A2 (estimated...
| File | emd_31536_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map with a B-factor of -122A2 (estimated automatically in relion), used during model building | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocessed map with a B-factor of -20A2, used...
| File | emd_31536_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map with a B-factor of -20A2, used during model building | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: one of the half map from refinement
| File | emd_31536_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | one of the half map from refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: one of the half map from refinement
| File | emd_31536_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | one of the half map from refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human FL mGluR5-5M with L-quisqualic acid and PAM VU0424465, agon...
| Entire | Name: Human FL mGluR5-5M with L-quisqualic acid and PAM VU0424465, agonist bound state |
|---|---|
| Components |
|
-Supramolecule #1: Human FL mGluR5-5M with L-quisqualic acid and PAM VU0424465, agon...
| Supramolecule | Name: Human FL mGluR5-5M with L-quisqualic acid and PAM VU0424465, agonist bound state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 194 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 5
| Macromolecule | Name: Metabotropic glutamate receptor 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.953977 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKHH HHHHHHHHLE VLFQGPQSSE RRVVAHMPGD IIIGALFSVH HQPTVDKVHE RKCGAVREQY GIQRVEAMLH TLERINSDP TLLPNITLGC EIRDSCWHSA VALEQSIEFI RDSLISSEEE EGLVRCVDGS SSSFRSKKPI VGVIGPGSSS V AIQVQNLL ...String: DYKDDDDKHH HHHHHHHHLE VLFQGPQSSE RRVVAHMPGD IIIGALFSVH HQPTVDKVHE RKCGAVREQY GIQRVEAMLH TLERINSDP TLLPNITLGC EIRDSCWHSA VALEQSIEFI RDSLISSEEE EGLVRCVDGS SSSFRSKKPI VGVIGPGSSS V AIQVQNLL QLFNIPQIAY SATSMDLSDK TLFKYFMRVV PSDAQQARAM VDIVKRYNWT YVSAVHTEGN YGESGMEAFK DM SAKEGIC IAHSYKIYSN AGEQSFDKLL KKLTSHLPKA RVVACFCEGM TVRGLLMAMR RLGLAGEFLL LGSDGWADRY DVT DGYQRE AVGGITIKLQ SPDVKWFDDY YLKLRPETNL RNPWFQEFWQ HRFQCRLEGF PQENSKYNKT CNSSLTLKTH HVQD SKMGF VINAIYSMAY GLHNMQMSLC PGYAGLCDAM KPIDGRKLLE SLMKTAFTGV SGDTILFDEN GDSPGRYEIM NFKEM GKDY FDYINVGSWD NGELKMDDDE VWSKKSNIIR SVCSEPCEKG QIKVIRKGEV SCCWTCTPCK ENEYVFDEYT CKACQL GSW PTDDLTGCDL IPVQYLRWGD PEPIAAVVFA CLGLLATLFV TVVFIIYRDT PVVKSSSREL CYIILAGICL GYLCTFC LI AKPKQIYCYL QRIGIGLSPA MSYSALVTKT NRIARILAGS KKKICTKKPR FMSACAQLVI AFILICIQLG IIVALFIM E PPDIMHDYPS IREVYLICNT TNLGVVAPLG YNGLLILACT FYAFKTRNVP ANFNEAKYIA FAMYTTCIIW LAFVPIYFG SNYKAITMCF SVSLSATVLL GCMFVPKVYI ILAKPERNVR SAFTTSTVVR MHVGDGKSSS AA UniProtKB: Metabotropic glutamate receptor 5 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID
| Macromolecule | Name: (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID type: ligand / ID: 3 / Number of copies: 2 / Formula: QUS |
|---|---|
| Molecular weight | Theoretical: 189.126 Da |
| Chemical component information | 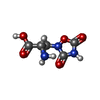 ChemComp-QUS: |
-Macromolecule #4: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.85 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force was 10, 3.5 seconds blotting time. | |||||||||
| Details | mGluR5 with L-quisqualic acid and ago PAM (VU0424464) purified in DDM/CHS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV Details: Imaging was done in EFTEM mode. Images were collected using beam shift in a series of 3x3 holes before stage shift. Active beam tilt compensation was turned on during the acquisition. |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 14604 / Average exposure time: 9.0 sec. / Average electron dose: 67.0 e/Å2 Details: Dose rate was 5.894 e/p/s. Total number of frames is 48 and each frame had a dose of 1.39 e/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 44500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Refinement was performed with jelly body restraints | ||||||||
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 113.4 | ||||||||
| Output model |  PDB-7fd8: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)