[English] 日本語
 Yorodumi
Yorodumi- EMDB-32356: Computational design of a potent Epstein-Barr virus fusion protei... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virology / Microbiology / Vaccine / EBV / IMMUNE SYSTEM | |||||||||
| Biological species |  Human gammaherpesvirus 4 (Epstein-Barr virus) Human gammaherpesvirus 4 (Epstein-Barr virus) | |||||||||
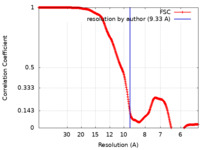
| Method | subtomogram averaging / cryo EM / Resolution: 9.33 Å | |||||||||
 Authors Authors | Sun C / Wang AJ / Fang XY / Gao YZ / Liu Z / Zeng MS | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2023 Journal: Cell Host Microbe / Year: 2023Title: A gB nanoparticle vaccine elicits a protective neutralizing antibody response against EBV Authors: Sun C / Kang YF / Fang XY / Liu YN / Bu GL / Wang AJ / Li Y / Zhu QY / Zhang H / Xie C / Kong XW / Peng YJ / Lin WJ / Zhou L / Chen XC / Lu ZZ / Xu HQ / Hong DC / Zhang X / Zhong L / Feng GK ...Authors: Sun C / Kang YF / Fang XY / Liu YN / Bu GL / Wang AJ / Li Y / Zhu QY / Zhang H / Xie C / Kong XW / Peng YJ / Lin WJ / Zhou L / Chen XC / Lu ZZ / Xu HQ / Hong DC / Zhang X / Zhong L / Feng GK / Zeng YX / Xu M / Zhong Q / Liu Z / Zeng MS | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32356.map.gz emd_32356.map.gz | 917.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32356-v30.xml emd-32356-v30.xml emd-32356.xml emd-32356.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32356_fsc.xml emd_32356_fsc.xml | 33.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_32356.png emd_32356.png | 37.6 KB | ||
| Filedesc metadata |  emd-32356.cif.gz emd-32356.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32356 http://ftp.pdbj.org/pub/emdb/structures/EMD-32356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32356 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32356.map.gz / Format: CCP4 / Size: 8.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32356.map.gz / Format: CCP4 / Size: 8.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.668 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : gB-I53-50 NP
| Entire | Name: gB-I53-50 NP |
|---|---|
| Components |
|
-Supramolecule #1: gB-I53-50 NP
| Supramolecule | Name: gB-I53-50 NP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human gammaherpesvirus 4 (Epstein-Barr virus) Human gammaherpesvirus 4 (Epstein-Barr virus) |
-Macromolecule #1: glycoprotein B
| Macromolecule | Name: glycoprotein B / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Human gammaherpesvirus 4 (Epstein-Barr virus) Human gammaherpesvirus 4 (Epstein-Barr virus) |
| Sequence | String: QTPEQPAPPA TTVQPTATRQ QTSFPFRVCE LSSHGDLFRF SSDIQCPSFG TRENHTEGLL MVFKDNIIPY SFKVRSYTKI VTNILIYNGH RADSVTNRHE EKFSVESYET DQMDTIYQCY NAVKMTKDGL TRVYVDRDGV NITVNLKPTG GLANGVRRYA SQTELYDAPG ...String: QTPEQPAPPA TTVQPTATRQ QTSFPFRVCE LSSHGDLFRF SSDIQCPSFG TRENHTEGLL MVFKDNIIPY SFKVRSYTKI VTNILIYNGH RADSVTNRHE EKFSVESYET DQMDTIYQCY NAVKMTKDGL TRVYVDRDGV NITVNLKPTG GLANGVRRYA SQTELYDAPG RVEATYRTRT TVNCLITDMM AKSNSPFDFF VTTTGQTVEM SPFYDGKNTE TFHERADSFH VRTNYKIVDY DNRGTNPQGE RRAFLDKGTY TLSWKLENRT AYCPLQHWQT FDSTIATETG KSIHFVTDEG TSSFVTNTTV GIELPDAFKC IEEQVNKTMH EKYEAVQDRY TKGQEAITYF ITSGGLLLAW LPLTPRSLAT VKNLTELTTP TSSPPSSPSP PAPPAARGST SAAVLRRRRR NAGNATTPVP PAAPGKSLGT LNNPATVQIQ FAYDSLRRQI NRMLGDLARA WCLEQKRQNM VLRELTKINP TTVMSSIYGK AVAAKRLGDV ISVSQCVPVN QATVTLRKSM RVPGSETMCY SRPLVSFSFI NDTKTYEGQL GTDNEIFLTK KMTEVCQATS QYYFQSGNEI HVYNDYHHFK TIELDGIATL QTFISLNTSL IENIDFASLE LYSRDEQRAS NVFDLEGIFR EYNFQAQNIA GLRKDLDNAV S |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 2.44 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 53000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)