-Search query
-Search result
Showing 1 - 50 of 1,105 items for (author: li & dg)

EMDB-41433: 
Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter

EMDB-41437: 
Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter

EMDB-41439: 
Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter

EMDB-41448: 
Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter

EMDB-41456: 
Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter

PDB-8to1: 
Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter

PDB-8to6: 
Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter

PDB-8to8: 
Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter

PDB-8toe: 
Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter

PDB-8tom: 
Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter

EMDB-40981: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP3

EMDB-40982: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody N289

EMDB-40822: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF1
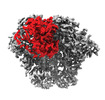
EMDB-40823: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF3

EMDB-40824: 
BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base4

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-41152: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation

PDB-8tc5: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation

EMDB-19576: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN

EMDB-19582: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : LM32Cs3H1 sKO STRAIN

EMDB-17216: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN

EMDB-18889: 
Mouse teneurin-3 compact dimer - A1B1 isoform

EMDB-18890: 
Mouse teneurin-3 non-compact subunit - A1B1 isoform

EMDB-18891: 
Mouse teneurin-3 non-compact subunit - A0B0 isoform

EMDB-19409: 
Mouse teneurin-3 compact dimer - A1B0 isoform

PDB-8r50: 
Mouse teneurin-3 compact dimer - A1B1 isoform

PDB-8r51: 
Mouse teneurin-3 non-compact subunit - A1B1 isoform

PDB-8r54: 
Mouse teneurin-3 non-compact subunit - A0B0 isoform

EMDB-41149: 
Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation

EMDB-41150: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation

PDB-8tc0: 
Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation

PDB-8tc1: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation
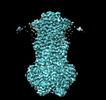
EMDB-40180: 
MsbA bound to cerastecin C

PDB-8gk7: 
MsbA bound to cerastecin C

EMDB-37736: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1)

EMDB-37737: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2)

EMDB-37739: 
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1
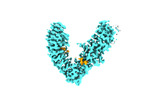
EMDB-37740: 
Local refinement of FEM1B bound with the C-degron of CCC89

EMDB-37742: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1)

EMDB-37743: 
cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2)
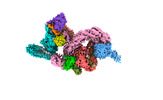
EMDB-37744: 
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1)

EMDB-37745: 
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2)

EMDB-37746: 
Local refinement of FEM1B bound with the C-degron of CUX1

PDB-8wqa: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1)

PDB-8wqb: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2)

PDB-8wqc: 
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1

PDB-8wqd: 
Local refinement of FEM1B bound with the C-degron of CCC89

PDB-8wqe: 
Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1)

PDB-8wqf: 
cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2)

PDB-8wqg: 
cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
