+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse teneurin-3 non-compact subunit - A1B1 isoform | |||||||||
 Map data Map data | Map of mouse teneurin-3 A1B1 isoform non-compact subunit | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic cell adhesion molecule / Homodimer / Cis-synaptic / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction ...regulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction / protein homodimerization activity / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
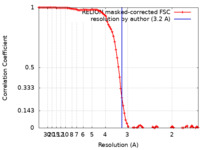
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Gogou C / Meijer DH | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition. Authors: Christos Gogou / J Wouter Beugelink / Cátia P Frias / Leanid Kresik / Natalia Jaroszynska / Uwe Drescher / Bert J C Janssen / Robert Hindges / Dimphna H Meijer /   Abstract: Neuronal network formation is facilitated by recognition between synaptic cell adhesion molecules at the cell surface. Alternative splicing of cell adhesion molecules provides additional specificity ...Neuronal network formation is facilitated by recognition between synaptic cell adhesion molecules at the cell surface. Alternative splicing of cell adhesion molecules provides additional specificity in forming neuronal connections. For the teneurin family of cell adhesion molecules, alternative splicing of the EGF-repeats and NHL domain controls synaptic protein-protein interactions. Here we present cryo-EM structures of the compact dimeric ectodomain of two teneurin-3 isoforms that harbour the splice insert in the EGF-repeats. This dimer is stabilised by an EGF8-ABD contact between subunits. Cryo-EM reconstructions of all four splice variants, together with SAXS and negative stain EM, reveal compacted dimers for each, with variant-specific dimeric arrangements. This results in specific trans-cellular interactions, as tested in cell clustering and stripe assays. The compact conformations provide a structural basis for teneurin homo- and heterophilic interactions. Altogether, our findings demonstrate how alternative splicing results in rearrangements of the dimeric subunits, influencing neuronal recognition and likely circuit wiring. #1:  Journal: BioRxiv Journal: BioRxivTitle: Mouse teneurin-3 non-compact subunit - A1B1 isoform Authors: Gogou C / Meijer DH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18890.map.gz emd_18890.map.gz | 62.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18890-v30.xml emd-18890-v30.xml emd-18890.xml emd-18890.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18890_fsc.xml emd_18890_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_18890.png emd_18890.png | 114.7 KB | ||
| Filedesc metadata |  emd-18890.cif.gz emd-18890.cif.gz | 8.1 KB | ||
| Others |  emd_18890_additional_1.map.gz emd_18890_additional_1.map.gz emd_18890_half_map_1.map.gz emd_18890_half_map_1.map.gz emd_18890_half_map_2.map.gz emd_18890_half_map_2.map.gz | 62.8 MB 52.1 MB 52.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18890 http://ftp.pdbj.org/pub/emdb/structures/EMD-18890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18890 | HTTPS FTP |
-Related structure data
| Related structure data |  8r51MC  8r50C  8r54C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18890.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18890.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of mouse teneurin-3 A1B1 isoform non-compact subunit | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.836 Å | ||||||||||||||||||||||||||||||||||||
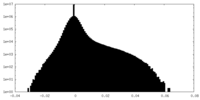
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map of mouse teneurin-3 A1B1 isoform non-compact subunit...
| File | emd_18890_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of mouse teneurin-3 A1B1 isoform non-compact subunit after focused classification and refinement on EGF8 | ||||||||||||
| Projections & Slices |
| ||||||||||||
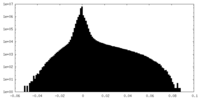
| Density Histograms |
-Half map: Half map of mouse teneurin-3 A1B1 isoform non-compact subunit
| File | emd_18890_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of mouse teneurin-3 A1B1 isoform non-compact subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
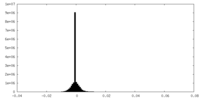
| Density Histograms |
-Half map: Half map of mouse teneurin-3 A1B1 isoform non-compact subunit
| File | emd_18890_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of mouse teneurin-3 A1B1 isoform non-compact subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
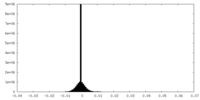
| Density Histograms |
- Sample components
Sample components
-Entire : mouse teneurin-3 A1B1 isoform ectodomain subunit
| Entire | Name: mouse teneurin-3 A1B1 isoform ectodomain subunit |
|---|---|
| Components |
|
-Supramolecule #1: mouse teneurin-3 A1B1 isoform ectodomain subunit
| Supramolecule | Name: mouse teneurin-3 A1B1 isoform ectodomain subunit / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 536.96 KDa |
-Macromolecule #1: Teneurin-3
| Macromolecule | Name: Teneurin-3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 268.72225 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARPLCTLLL LMATLAGALA GSHHHHHHGS QTENDTFENG KVNSDTVPTN TVSLPSGDNG KLGGFTHENN TIDSGELDIG RRAIQEVPP GIFWRSQLFI DQPQFLKFNI SLQKDALIGV YGRKGLPPSH TQYDFVELLD GSRLIAREQR NLVESERAGR Q ARSVSLHE ...String: MARPLCTLLL LMATLAGALA GSHHHHHHGS QTENDTFENG KVNSDTVPTN TVSLPSGDNG KLGGFTHENN TIDSGELDIG RRAIQEVPP GIFWRSQLFI DQPQFLKFNI SLQKDALIGV YGRKGLPPSH TQYDFVELLD GSRLIAREQR NLVESERAGR Q ARSVSLHE AGFIQYLDSG IWHLAFYNDG KNPEQVSFNT IVIESVVECP RNCHGNGECV SGTCHCFPGF LGPDCSRAAC PV LCSGNGQ YSKGRCLCFS GWKGTECDVP TTQCIDPQCG GRGICIMGSC ACNSGYKGEN CEEADCLDPG CSNHGVCIHG ECH CNPGWG GSNCEILKTM CADQCSGHGT YLQESGSCTC DPNWTGPDCS NEICSVDCGS HGVCMGGSCR CEEGWTGPAC NQRA CHPRC AEHGTCKDGK CECSQGWNGE HCTIAHYLDK IVKEGCPGLC NSNGRCTLDQ NGWHCVCQPG WRGAGCDVAM ETLCT DSKD NEGDGLIDCM DPDCCLQSSC QNQPYCRGLP DPQDIISQSL QTPSQQAAKS FYDRISFLIG SDSTHVLPGE SPFNKS LAS VIRGQVLTAD GTPLIGVNVS FLHYSEYGYT ITRQDGMFDL VANGGASLTL VFERSPFLTQ YHTVWIPWNV FYVMDTL VM KKEENDIPSC DLSGFVRPSP IIVSSPLSTF FRSSPEDSPI IPETQVLHEE TTIPGTDLKL SYLSSRAAGY KSVLKITM T QAVIPFNLMK VHLMVAVVGR LFQKWFPASP NLAYTFIWDK TDAYNQKVYG LSEAVVSVGY EYESCLDLTL WEKRTAVLQ GYELDASNMG GWTLDKHHVL DVQNGILYKG NGENQFISQQ PPVVSSIMGN GRRRSISCPS CNGQADGNKL LAPVALACGI DGSLYVGDF NYVRRIFPSG NVTSVLELRN KDFRHSSNPA HRYYLATDPV TGDLYVSDTN TRRIYRPKSL TGAKDLTKNA E VVAGTGEQ CLPFDEARCG DGGKAVEATL MSPKGMAIDK NGLIYFVDGT MIRKVDQNGI ISTLLGSNDL TSARPLTCDT SM HISQVRL EWPTDLAINP MDNSIYVLDN NVVLQITENR QVRIAAGRPM HCQVPGVEYP VGKHAVQTTL ESATAIAVSY SGV LYITET DEKKINRIRQ VTTDGEISLV AGIPSECDCK NDANCDCYQS GDGYAKDAKL NAPSSLAASP DGTLYIADLG NIRI RAVSK NKPLLNSMNF YEVASPTDQE LYIFDINGTH QYTVSLVTGD YLYNFSYSND NDVTAVTDSN GNTLRIRRDP NRMPV RVVS PDNQVIWLTI GTNGCLKSMT AQGLELVLFT YHGNSGLLAT KSDETGWTTF FDYDSEGRLT NVTFPTGVVT NLHGDM DKA ITVDIESSSR EEDVSITSNL SSIDSFYTMV QDQLRNSYQI GYDGSLRIFY ASGLDSHYQT EPHVLAGTAN PTVAKRN MT LPGENGQNLV EWRFRKEQAQ GKVNVFGRKL RVNGRNLLSV DFDRTTKTEK IYDDHRKFLL RIAYDTSGHP TLWLPSSK L MAVNVTYSST GQIASIQRGT TSEKVDYDSQ GRIVSRVFAD GKTWSYTYLE KSMVLLLHSQ RQYIFEYDMW DRLSAITMP SVARHTMQTI RSIGYYRNIY NPPESNASII TDYNEEGLLL QTAFLGTSRR VLFKYRRQTR LSEILYDSTR VSFTYDETAG VLKTVNLQS DGFICTIRYR QIGPLIDRQI FRFSEDGMVN ARFDYSYDNS FRVTSMQGVI NETPLPIDLY QFDDISGKVE Q FGKFGVIY YDINQIISTA VMTYTKHFDA HGRIKEIQYE IFRSLMYWIT IQYDNMGRVT KREIKIGPFA NTTKYAYEYD VD GQLQTVY LNEKIMWRYN YDLNGNLHLL NPSSSARLTP LRYDLRDRIT RLGDVQYRLD EDGFLRQRGT EIFEYSSKGL LTR VYSKGS GWTVIYRYDG LGRRVSSKTS LGQHLQFFYA DLTYPTRITH VYNHSSSEIT SLYYDLQGHL FAMEISSGDE FYIA SDNTG TPLAVFSSNG LMLKQIQYTA YGEIYFDSNV DFQLVIGFHG GLYDPLTKLI HFGERDYDIL AGRWTTPDIE IWKRI GKDP APFNLYMFRN NNPASKIHDV KDYITDVNSW LVTFGFHLHN AIPGFPVPKF DLTEPSYELV KSQQWEDVPP IFGVQQ QVA RQAKAFLSLG KMAEVQVSRR KAGAEQSWLW FATVKSLIGK GVMLAVSQGR VQTNVLNIAN EDCIKVAAVL NNAFYLE NL HFTIEGKDTH YFIKTTTPES DLGTLRLTSG RKALENGINV TVSQSTTVVN GRTRRFADVE MQFGALALHV RYGMTLDE E KARILEQARQ RALARAWARE QQRVRDGEEG ARLWTEGEKR QLLSAGKVQG YDGYYVLSVE QYPELADSAN NIQFLRQSE IGKRAAA UniProtKB: Teneurin-3 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 20 mM HEPES, 150 mM NaCl, 2mM CaCl2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 22 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)