+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse teneurin-3 non-compact subunit - A0B0 isoform | |||||||||
 Map data Map data | Map of the mouse teneurin-3 A0B0 isoform ectodomain non-compact subunit | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Synaptic cell adhesion molecule / Homodimer / Cis-synaptic / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction ...regulation of homophilic cell adhesion / synaptic membrane adhesion / homophilic cell-cell adhesion / presynaptic active zone membrane / positive regulation of neuron projection development / cell differentiation / protein heterodimerization activity / axon / glutamatergic synapse / signal transduction / protein homodimerization activity / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Gogou C / Meijer DH | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition. Authors: Christos Gogou / J Wouter Beugelink / Cátia P Frias / Leanid Kresik / Natalia Jaroszynska / Uwe Drescher / Bert J C Janssen / Robert Hindges / Dimphna H Meijer /   Abstract: Neuronal network formation is facilitated by recognition between synaptic cell adhesion molecules at the cell surface. Alternative splicing of cell adhesion molecules provides additional specificity ...Neuronal network formation is facilitated by recognition between synaptic cell adhesion molecules at the cell surface. Alternative splicing of cell adhesion molecules provides additional specificity in forming neuronal connections. For the teneurin family of cell adhesion molecules, alternative splicing of the EGF-repeats and NHL domain controls synaptic protein-protein interactions. Here we present cryo-EM structures of the compact dimeric ectodomain of two teneurin-3 isoforms that harbour the splice insert in the EGF-repeats. This dimer is stabilised by an EGF8-ABD contact between subunits. Cryo-EM reconstructions of all four splice variants, together with SAXS and negative stain EM, reveal compacted dimers for each, with variant-specific dimeric arrangements. This results in specific trans-cellular interactions, as tested in cell clustering and stripe assays. The compact conformations provide a structural basis for teneurin homo- and heterophilic interactions. Altogether, our findings demonstrate how alternative splicing results in rearrangements of the dimeric subunits, influencing neuronal recognition and likely circuit wiring. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18891.map.gz emd_18891.map.gz | 49.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18891-v30.xml emd-18891-v30.xml emd-18891.xml emd-18891.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18891_fsc.xml emd_18891_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18891.png emd_18891.png | 112.9 KB | ||
| Filedesc metadata |  emd-18891.cif.gz emd-18891.cif.gz | 7.9 KB | ||
| Others |  emd_18891_half_map_1.map.gz emd_18891_half_map_1.map.gz emd_18891_half_map_2.map.gz emd_18891_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18891 http://ftp.pdbj.org/pub/emdb/structures/EMD-18891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18891 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18891 | HTTPS FTP |
-Related structure data
| Related structure data |  8r54MC  8r50C  8r51C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18891.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18891.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the mouse teneurin-3 A0B0 isoform ectodomain non-compact subunit | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.859 Å | ||||||||||||||||||||||||||||||||||||
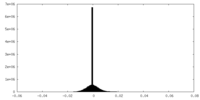
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of the mouse teneurin-3 A0B0 isoform...
| File | emd_18891_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the mouse teneurin-3 A0B0 isoform ectodomain non-compact subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
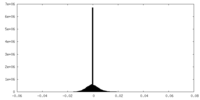
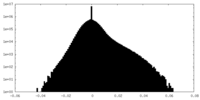
| Density Histograms |
-Half map: Half map of the mouse teneurin-3 A0B0 isoform...
| File | emd_18891_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the mouse teneurin-3 A0B0 isoform ectodomain non-compact subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse teneurin-3 A0B0 isoform ectodomain subunit
| Entire | Name: Mouse teneurin-3 A0B0 isoform ectodomain subunit |
|---|---|
| Components |
|
-Supramolecule #1: Mouse teneurin-3 A0B0 isoform ectodomain subunit
| Supramolecule | Name: Mouse teneurin-3 A0B0 isoform ectodomain subunit / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 532.92 KDa |
-Macromolecule #1: Teneurin-3
| Macromolecule | Name: Teneurin-3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 266.692844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARPLCTLLL LMATLAGALA GSHHHHHHGS QTENDTFENG KVNSDTVPTN TVSLPSGDNG KLGGFTHENN TIDSGELDIG RRAIQEVPP GIFWRSQLFI DQPQFLKFNI SLQKDALIGV YGRKGLPPSH TQYDFVELLD GSRLIAREQR NLVESERAGR Q ARSVSLHE ...String: MARPLCTLLL LMATLAGALA GSHHHHHHGS QTENDTFENG KVNSDTVPTN TVSLPSGDNG KLGGFTHENN TIDSGELDIG RRAIQEVPP GIFWRSQLFI DQPQFLKFNI SLQKDALIGV YGRKGLPPSH TQYDFVELLD GSRLIAREQR NLVESERAGR Q ARSVSLHE AGFIQYLDSG IWHLAFYNDG KNPEQVSFNT IVIESVVECP RNCHGNGECV SGTCHCFPGF LGPDCSRAAC PV LCSGNGQ YSKGRCLCFS GWKGTECDVP TTQCIDPQCG GRGICIMGSC ACNSGYKGEN CEEADCLDPG CSNHGVCIHG ECH CNPGWG GSNCEILKTM CADQCSGHGT YLQESGSCTC DPNWTGPDCS NEICSVDCGS HGVCMGGSCR CEEGWTGPAC NQRA CHPRC AEHGTCKDGK CECSQGWNGE HCTIEGCPGL CNSNGRCTLD QNGWHCVCQP GWRGAGCDVA METLCTDSKD NEGDG LIDC MDPDCCLQSS CQNQPYCRGL PDPQDIISQS LQTPSQQAAK SFYDRISFLI GSDSTHVLPG ESPFNKSLAS VIRGQV LTA DGTPLIGVNV SFLHYSEYGY TITRQDGMFD LVANGGASLT LVFERSPFLT QYHTVWIPWN VFYVMDTLVM KKEENDI PS CDLSGFVRPS PIIVSSPLST FFRSSPEDSP IIPETQVLHE ETTIPGTDLK LSYLSSRAAG YKSVLKITMT QAVIPFNL M KVHLMVAVVG RLFQKWFPAS PNLAYTFIWD KTDAYNQKVY GLSEAVVSVG YEYESCLDLT LWEKRTAVLQ GYELDASNM GGWTLDKHHV LDVQNGILYK GNGENQFISQ QPPVVSSIMG NGRRRSISCP SCNGQADGNK LLAPVALACG IDGSLYVGDF NYVRRIFPS GNVTSVLELS SNPAHRYYLA TDPVTGDLYV SDTNTRRIYR PKSLTGAKDL TKNAEVVAGT GEQCLPFDEA R CGDGGKAV EATLMSPKGM AIDKNGLIYF VDGTMIRKVD QNGIISTLLG SNDLTSARPL TCDTSMHISQ VRLEWPTDLA IN PMDNSIY VLDNNVVLQI TENRQVRIAA GRPMHCQVPG VEYPVGKHAV QTTLESATAI AVSYSGVLYI TETDEKKINR IRQ VTTDGE ISLVAGIPSE CDCKNDANCD CYQSGDGYAK DAKLNAPSSL AASPDGTLYI ADLGNIRIRA VSKNKPLLNS MNFY EVASP TDQELYIFDI NGTHQYTVSL VTGDYLYNFS YSNDNDVTAV TDSNGNTLRI RRDPNRMPVR VVSPDNQVIW LTIGT NGCL KSMTAQGLEL VLFTYHGNSG LLATKSDETG WTTFFDYDSE GRLTNVTFPT GVVTNLHGDM DKAITVDIES SSREED VSI TSNLSSIDSF YTMVQDQLRN SYQIGYDGSL RIFYASGLDS HYQTEPHVLA GTANPTVAKR NMTLPGENGQ NLVEWRF RK EQAQGKVNVF GRKLRVNGRN LLSVDFDRTT KTEKIYDDHR KFLLRIAYDT SGHPTLWLPS SKLMAVNVTY SSTGQIAS I QRGTTSEKVD YDSQGRIVSR VFADGKTWSY TYLEKSMVLL LHSQRQYIFE YDMWDRLSAI TMPSVARHTM QTIRSIGYY RNIYNPPESN ASIITDYNEE GLLLQTAFLG TSRRVLFKYR RQTRLSEILY DSTRVSFTYD ETAGVLKTVN LQSDGFICTI RYRQIGPLI DRQIFRFSED GMVNARFDYS YDNSFRVTSM QGVINETPLP IDLYQFDDIS GKVEQFGKFG VIYYDINQII S TAVMTYTK HFDAHGRIKE IQYEIFRSLM YWITIQYDNM GRVTKREIKI GPFANTTKYA YEYDVDGQLQ TVYLNEKIMW RY NYDLNGN LHLLNPSSSA RLTPLRYDLR DRITRLGDVQ YRLDEDGFLR QRGTEIFEYS SKGLLTRVYS KGSGWTVIYR YDG LGRRVS SKTSLGQHLQ FFYADLTYPT RITHVYNHSS SEITSLYYDL QGHLFAMEIS SGDEFYIASD NTGTPLAVFS SNGL MLKQI QYTAYGEIYF DSNVDFQLVI GFHGGLYDPL TKLIHFGERD YDILAGRWTT PDIEIWKRIG KDPAPFNLYM FRNNN PASK IHDVKDYITD VNSWLVTFGF HLHNAIPGFP VPKFDLTEPS YELVKSQQWE DVPPIFGVQQ QVARQAKAFL SLGKMA EVQ VSRRKAGAEQ SWLWFATVKS LIGKGVMLAV SQGRVQTNVL NIANEDCIKV AAVLNNAFYL ENLHFTIEGK DTHYFIK TT TPESDLGTLR LTSGRKALEN GINVTVSQST TVVNGRTRRF ADVEMQFGAL ALHVRYGMTL DEEKARILEQ ARQRALAR A WAREQQRVRD GEEGARLWTE GEKRQLLSAG KVQGYDGYYV LSVEQYPELA DSANNIQFLR QSEIGKRAAA UniProtKB: Teneurin-3 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 20 mM HEPES, 150 mM NaCl, 2mM CaCl2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)