[English] 日本語
 Yorodumi
Yorodumi- EMDB-40981: BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP3 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP3 | ||||||||||||
 Map data Map data | Fp-3 post process | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / Env / NHP / Antibody / Fusion Peptide / Polyclonal / VIRAL PROTEIN | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | ||||||||||||
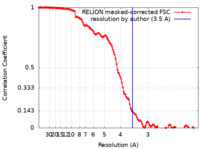
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Pratap PP / Antansijevic A / Ozorowski G / Ward AB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2024 Journal: NPJ Vaccines / Year: 2024Title: Priming antibody responses to the fusion peptide in rhesus macaques. Authors: Christopher A Cottrell / Payal P Pratap / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A Enemuo / Aleksandar Antanasijevic / Gabriel Ozorowski / Leigh M Sewall / Hongmei Gao / Joel D ...Authors: Christopher A Cottrell / Payal P Pratap / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A Enemuo / Aleksandar Antanasijevic / Gabriel Ozorowski / Leigh M Sewall / Hongmei Gao / Joel D Allen / Bartek Nogal / Murillo Silva / Jinal Bhiman / Matthias Pauthner / Darrell J Irvine / David Montefiori / Max Crispin / Dennis R Burton / Guido Silvestri / Shane Crotty / Andrew B Ward /   Abstract: Immunodominance of antibodies targeting non-neutralizing epitopes and the high level of somatic hypermutation within germinal centers (GCs) required for most HIV broadly neutralizing antibodies ...Immunodominance of antibodies targeting non-neutralizing epitopes and the high level of somatic hypermutation within germinal centers (GCs) required for most HIV broadly neutralizing antibodies (bnAbs) are major impediments to the development of an effective HIV vaccine. Rational protein vaccine design and non-conventional immunization strategies are potential avenues to overcome these hurdles. Here, we report using implantable osmotic pumps to continuously deliver a series of epitope-targeted immunogens to rhesus macaques over the course of six months to prime and elicit antibody responses against the conserved fusion peptide (FP). GC responses and antibody specificities were tracked longitudinally using lymph node fine-needle aspirates and electron microscopy polyclonal epitope mapping (EMPEM), respectively, to show antibody responses to the FP/N611 glycan hole region were primed, although exhibited limited neutralization breadth. Application of cryoEMPEM delineated key residues for on-target and off-target responses that can drive the next round of structure-based vaccine design. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40981.map.gz emd_40981.map.gz | 166.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40981-v30.xml emd-40981-v30.xml emd-40981.xml emd-40981.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40981_fsc.xml emd_40981_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_40981.png emd_40981.png | 168.9 KB | ||
| Masks |  emd_40981_msk_1.map emd_40981_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40981.cif.gz emd-40981.cif.gz | 7.8 KB | ||
| Others |  emd_40981_half_map_1.map.gz emd_40981_half_map_1.map.gz emd_40981_half_map_2.map.gz emd_40981_half_map_2.map.gz | 141 MB 140.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40981 http://ftp.pdbj.org/pub/emdb/structures/EMD-40981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40981 | HTTPS FTP |
-Related structure data
| Related structure data |  8t2eMC  8swvC  8swwC  8swxC  8t2fC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40981.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40981.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fp-3 post process | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.031 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40981_msk_1.map emd_40981_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_40981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_40981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 Boost 2 in complex with NHP Polyclonal Antibody FP3
| Entire | Name: BG505 Boost 2 in complex with NHP Polyclonal Antibody FP3 |
|---|---|
| Components |
|
-Supramolecule #1: BG505 Boost 2 in complex with NHP Polyclonal Antibody FP3
| Supramolecule | Name: BG505 Boost 2 in complex with NHP Polyclonal Antibody FP3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 470 KDa |
-Supramolecule #2: Surface protein gp120, Transmembrane protein gp41
| Supramolecule | Name: Surface protein gp120, Transmembrane protein gp41 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Supramolecule #3: FP3 Heavy Chain, FP3 Light Chain
| Supramolecule | Name: FP3 Heavy Chain, FP3 Light Chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Surface protein gp120
| Macromolecule | Name: Surface protein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 58.090172 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETKK HNVWATHCCV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETKK HNVWATHCCV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK VYSLFYRLDV VQINENQGNR SNNSNKEYRL INCNTSAITQ ACPKVSFEPI PIHYCAPAGF AILKCKDKKF NG TGPCTNV STVQCTHGIK PVVSTQLLLN GSLAEEEVII RSENITNNAK NILVQLNESV QINCTRPNNN TRKSIRIGPG QWF YATGDI IGDIRQAHCN VSKATWNETL GKVVKQLRKH FGNNTIIRFA NSSGGDLEVT THSFNCGGEF FYCNTSGLFN STWI SNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRD NWRS ELYKYKVVKI EPLGVAPTRC KRRVVGRRRR RR |
-Macromolecule #2: Transmembrane protein gp41
| Macromolecule | Name: Transmembrane protein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.192574 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEC QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSTWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #3: FP3 Heavy Chain
| Macromolecule | Name: FP3 Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.081651 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) |
-Macromolecule #4: FP3 Light Chain
| Macromolecule | Name: FP3 Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.528504 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK) |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 50 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7339 / Average electron dose: 41.3591843915 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)