-Search query
-Search result
Showing 1 - 50 of 65 items for (author: lee & lj)

EMDB-28850: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab

EMDB-28851: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-10 Fab

EMDB-28852: 
SARS-CoV-2 Omicron 6P S + COVA309-35 Fab
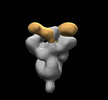
EMDB-28853: 
SARS-CoV-2 Gamma 6P Mut7 + S COVA309-38 Fab

EMDB-28755: 
Human tRNA Splicing Endonuclease Complex bound to 2'F-tRNA-Arg
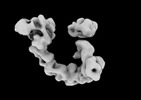
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2

EMDB-26856: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG

EMDB-26831: 
Human Rix1 sub-complex scaffold
EMDB-13586: 
Cryo-electron tomography of ASC signalling sites in pyroptotic cells (2)

EMDB-25474: 
CryoEM structure of the N-Terminal deleted Rix7 AAA-ATPase

EMDB-25582: 
CryoEM structure of the crosslinked Rix7 AAA-ATPase

EMDB-25659: 
CryoEM structure of the Rix7 D2 Walker B mutant

EMDB-23708: 
ATP-bound AMP-activated protein kinase

EMDB-24642: 
SARS-CoV-2 Spike bound to Fab PDI 210

EMDB-24643: 
SARS-CoV-2 Spike bound to Fab PDI 96

EMDB-24644: 
SARS-CoV-2 Spike bound to Fab PDI 215

EMDB-24645: 
SARS-CoV-2 Spike bound to Fab WCSL 119

EMDB-24646: 
SARS-CoV-2 Spike bound to Fab WCSL 129

EMDB-24647: 
SARS-CoV-2 Spike bound to Fab PDI 93

EMDB-24648: 
SARS-CoV-2 Spike bound to Fab PDI 222

EMDB-24649: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222

EMDB-22336: 
Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody

EMDB-22337: 
Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody

EMDB-24137: 
SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state

EMDB-24101: 
SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state

EMDB-23749: 
Human Cholecystokinin 1 receptor (CCK1R) Gs complex

EMDB-23750: 
Human Cholecystokinin 1 receptor (CCK1R) Gq chimera (mGsqi) complex

EMDB-12286: 
Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B

PDB-7ndg: 
Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B

EMDB-23042: 
Chimeric flavivirus between Binjari virus and Dengue virus serotype-2

EMDB-23043: 
Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus

EMDB-23044: 
Structure of West Nile virus (Kunjin)

EMDB-23045: 
Chimeric flavivirus between Binjari virus and Murray Valley encephalitis virus

EMDB-22610: 
Nucleotide bound SARS-CoV-2 Nsp15

EMDB-22611: 
SARS-CoV-2 wt-Nsp15 APO-state

EMDB-22612: 
SARS-CoV-2 Nsp15 H235A variant APO-state, dataset ii

EMDB-22613: 
SARS-CoV-2 Nsp15 H235A APO-state, dataset i

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b

EMDB-21683: 
Human secretin receptor Gs complex

EMDB-21972: 
Human secretin receptor Gs complex

EMDB-20417: 
Cryo-EM reconstruction of the mature chimeric BinJV/ZIKV-prME virion

EMDB-20438: 
Cryo-EM reconstruction of the mature chimeric BinJV/ZIKV-prME virion in complex with Fab C8

EMDB-20439: 
Cryo-EM reconstruction of the immature chimeric BinJV/ZIKV-prME virion

EMDB-9757: 
pCBH ParM filament

EMDB-9758: 
pCBH ParM filament

EMDB-0350: 
Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus

EMDB-7092: 
Cryo-EM structure of human insulin degrading enzyme in complex with insulin

EMDB-7090: 
Cryo-EM structure of human insulin degrading enzyme

EMDB-7091: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
