-Search query
-Search result
Showing 1 - 50 of 1,979 items for (author: kim & k)

EMDB-42676: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

EMDB-42999: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

PDB-8uwl: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

PDB-8v6u: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

EMDB-36271: 
Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer

PDB-8jhk: 
Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer

EMDB-41248: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

EMDB-41249: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

PDB-8th3: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

PDB-8th4: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

EMDB-36907: 
Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris

PDB-8k5o: 
Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris

EMDB-18658: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex

PDB-8qu9: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex
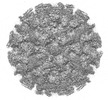
EMDB-36223: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp

PDB-8jg5: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp

EMDB-35866: 
PKR and NS1 complex

PDB-8izn: 
Structural study of Interferon-induced, double-stranded RNA-activated protein kinase (PKR) and Non-structural protein 1 (NS1) complex
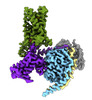
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
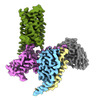
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)

PDB-8vy7: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant

PDB-8vy9: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane

EMDB-37386: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP

EMDB-37387: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC1

EMDB-37388: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2

PDB-8w9z: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP

PDB-8wa0: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC1

PDB-8wa1: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2

EMDB-41888: 
Structure of Apo CXCR4/Gi complex

EMDB-41889: 
Structure of CXCL12-bound CXCR4/Gi complex

EMDB-41890: 
Structure of AMD3100-bound CXCR4/Gi complex

EMDB-41891: 
Structure of REGN7663 Fab-bound CXCR4/Gi complex

EMDB-41892: 
Structure of REGN7663-Fab bound CXCR4

EMDB-41893: 
Structure of trimeric CXCR4 in complex with REGN7663 Fab

EMDB-41894: 
Structure of tetrameric CXCR4 in complex with REGN7663 Fab

EMDB-19789: 
Flexible reconstruction of the yeast U4/U6.U5 tri-snRNP (EMPIAR-10073) using DynaMight

EMDB-19791: 
Flexible reconstruction of a pre-catalytic spliceosome (EMPIAR-10180) using DynaMight

EMDB-19794: 
Flexible reconstruction of the yeast inner kinetochore bound to a CENP-A nucleosome (EMPIAR-11890)
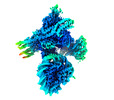
EMDB-19799: 
Flexible reconstruction of CBF1-CCAN bound to a centromeric CENP-A nucleosome (EMPIAR-11910)
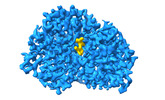
EMDB-39117: 
Cryo-EM structure of Maltose Binding Protein

PDB-8ybe: 
Cryo-EM structure of Maltose Binding Protein
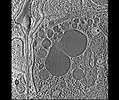
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
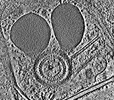
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
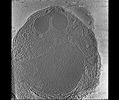
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite

EMDB-37465: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density

EMDB-37466: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
