+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CXCL12-bound CXCR4/Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / chemokine receptor / chemokine / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtelencephalon cell migration / chemokine (C-X-C motif) ligand 12 signaling pathway / C-X-C motif chemokine 12 receptor activity / response to ultrasound / negative regulation of leukocyte tethering or rolling / positive regulation of macrophage migration inhibitory factor signaling pathway / regulation of actin polymerization or depolymerization / chemokine receptor binding / myosin light chain binding / CXCL12-activated CXCR4 signaling pathway ...telencephalon cell migration / chemokine (C-X-C motif) ligand 12 signaling pathway / C-X-C motif chemokine 12 receptor activity / response to ultrasound / negative regulation of leukocyte tethering or rolling / positive regulation of macrophage migration inhibitory factor signaling pathway / regulation of actin polymerization or depolymerization / chemokine receptor binding / myosin light chain binding / CXCL12-activated CXCR4 signaling pathway / Specification of primordial germ cells / myelin maintenance / CXCR chemokine receptor binding / C-X-C chemokine receptor activity / positive regulation of axon extension involved in axon guidance / positive regulation of vasculature development / Signaling by ROBO receptors / regulation of chemotaxis / induction of positive chemotaxis / Formation of definitive endoderm / integrin activation / positive regulation of dopamine secretion / negative regulation of dendritic cell apoptotic process / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage / C-C chemokine receptor activity / cellular response to chemokine / Developmental Lineage of Pancreatic Acinar Cells / C-C chemokine binding / positive regulation of monocyte chemotaxis / chemokine activity / anchoring junction / Chemokine receptors bind chemokines / blood circulation / dendritic cell chemotaxis / chemokine-mediated signaling pathway / positive regulation of calcium ion import / cellular response to cytokine stimulus / detection of temperature stimulus involved in sensory perception of pain / cell leading edge / positive regulation of oligodendrocyte differentiation / animal organ regeneration / Binding and entry of HIV virion / detection of mechanical stimulus involved in sensory perception of pain / Nuclear signaling by ERBB4 / positive regulation of T cell migration / regulation of cell adhesion / adenylate cyclase inhibitor activity / coreceptor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / positive regulation of endothelial cell proliferation / response to prostaglandin E / D2 dopamine receptor binding / neurogenesis / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / positive regulation of neuron differentiation / positive regulation of cell adhesion / cellular response to forskolin / axon guidance / regulation of mitotic spindle organization / ubiquitin binding / adult locomotory behavior / cell chemotaxis / growth factor activity / Regulation of insulin secretion / calcium-mediated signaling / defense response / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / brain development / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / integrin binding / extracellular matrix / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to virus / centriolar satellite / G-protein beta/gamma-subunit complex binding / neuron migration / Olfactory Signaling Pathway / chemotaxis / Activation of the phototransduction cascade / intracellular calcium ion homeostasis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Saotome K / McGoldrick LL / Franklin MC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural insights into CXCR4 modulation and oligomerization. Authors: Kei Saotome / Luke L McGoldrick / Jo-Hao Ho / Trudy F Ramlall / Sweta Shah / Michael J Moore / Jee Hae Kim / Raymond Leidich / William C Olson / Matthew C Franklin /  Abstract: Activation of the chemokine receptor CXCR4 by its chemokine ligand CXCL12 regulates diverse cellular processes. Previously reported crystal structures of CXCR4 revealed the architecture of an ...Activation of the chemokine receptor CXCR4 by its chemokine ligand CXCL12 regulates diverse cellular processes. Previously reported crystal structures of CXCR4 revealed the architecture of an inactive, homodimeric receptor. However, many structural aspects of CXCR4 remain poorly understood. Here, we use cryo-electron microscopy to investigate various modes of human CXCR4 regulation. CXCL12 activates CXCR4 by inserting its N terminus deep into the CXCR4 orthosteric pocket. The binding of US Food and Drug Administration-approved antagonist AMD3100 is stabilized by electrostatic interactions with acidic residues in the seven-transmembrane-helix bundle. A potent antibody blocker, REGN7663, binds across the extracellular face of CXCR4 and inserts its complementarity-determining region H3 loop into the orthosteric pocket. Trimeric and tetrameric structures of CXCR4 reveal modes of G-protein-coupled receptor oligomerization. We show that CXCR4 adopts distinct subunit conformations in trimeric and tetrameric assemblies, highlighting how oligomerization could allosterically regulate chemokine receptor function. #1:  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Structural insights into CXCR4 modulation and oligomerization Authors: Saotome K / McGoldrick LL / Ho J / Ramlall T / Shah S / Moore MJ / Kim JH / Leidich R / Olson WC / Franklin MC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41889.map.gz emd_41889.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41889-v30.xml emd-41889-v30.xml emd-41889.xml emd-41889.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41889.png emd_41889.png | 99.7 KB | ||
| Filedesc metadata |  emd-41889.cif.gz emd-41889.cif.gz | 7.2 KB | ||
| Others |  emd_41889_half_map_1.map.gz emd_41889_half_map_1.map.gz emd_41889_half_map_2.map.gz emd_41889_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41889 http://ftp.pdbj.org/pub/emdb/structures/EMD-41889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41889 | HTTPS FTP |
-Related structure data
| Related structure data |  8u4oMC  8u4nC  8u4pC  8u4qC  8u4rC  8u4sC  8u4tC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41889.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41889.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
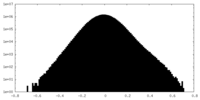
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41889_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
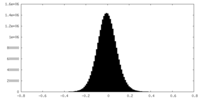
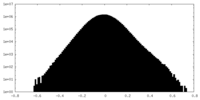
| Density Histograms |
-Half map: #2
| File | emd_41889_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
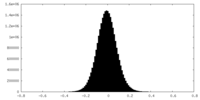
| Density Histograms |
- Sample components
Sample components
-Entire : CXCL12-bound CXCR4/Gi complex
| Entire | Name: CXCL12-bound CXCR4/Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: CXCL12-bound CXCR4/Gi complex
| Supramolecule | Name: CXCL12-bound CXCR4/Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: C-X-C chemokine receptor type 4
| Macromolecule | Name: C-X-C chemokine receptor type 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.063609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFAGAPE GISIYTSDNY TEEMGSGDYD SMKEPCFREE NANFNKIFLP TIYSIIFLTG IVGNGLVILV MGYQKKLRS MTDKYRLHLS VADLLFVITL PFWAVDAVAN WYFGNFLCKA VHVIYTVSLY SSVLILAFIS LDRYLAIVHA T NSQRPRKL ...String: MKTIIALSYI FCLVFAGAPE GISIYTSDNY TEEMGSGDYD SMKEPCFREE NANFNKIFLP TIYSIIFLTG IVGNGLVILV MGYQKKLRS MTDKYRLHLS VADLLFVITL PFWAVDAVAN WYFGNFLCKA VHVIYTVSLY SSVLILAFIS LDRYLAIVHA T NSQRPRKL LAEKVVYVGV WIPALLLTIP DFIFANVSEA DDRYICDRFY PNDLWVVVFQ FQHIMVGLIL PGIVILSCYC II ISKLSHS KGHQKRKALK TTVILILAFF ACWLPYYIGI SIDSFILLEI IKQGCEFENT VHKWISITEA LAFFHCCLNP ILY AFLGAK FKTSAQHALT SVSRGSSLKI LSKGKRGGHS SVSTESESSS FHSSGRPLEV LFQGPGGGGS VSKGEELFTG VVPI LVELD GDVNGHKFSV SGEGEGDATY GKLTLKFICT TGKLPVPWPT LVTTLTYGVQ CFSRYPDHMK QHDFFKSAMP EGYVQ ERTI FFKDDGNYKT RAEVKFEGDT LVNRIELKGI DFKEDGNILG HKLEYNYNSH NVYIMADKQK NGIKVNFKIR HNIEDG SVQ LADHYQQNTP IGDGPVLLPD NHYLSTQSKL SKDPNEKRDH MVLLEFVTAA GITLGMDELY KDYKDDDDK UniProtKB: C-X-C chemokine receptor type 4 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.591312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGGG GSGCTLSAED KAAVERSKMI DRNLREDGEK AAREVKLLLL GAGESGKCTI VKQMKIIHEA GYSEEECKQY KAVVYSNTI QSIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL N DSAAYYLN ...String: MHHHHHHGGG GSGCTLSAED KAAVERSKMI DRNLREDGEK AAREVKLLLL GAGESGKCTI VKQMKIIHEA GYSEEECKQY KAVVYSNTI QSIIAIIRAM GRLKIDFGDS ARADDARQLF VLAGAAEEGF MTAELAGVIK RLWKDSGVQA CFNRSREYQL N DSAAYYLN DLDRIAQPNY IPTQQDVLRT RVKTTGIVET HFTFKDLHFK MFDVTAQRSE RKKWIHCFEG VTAIIFCVAL SD YDLVLAE DEEMNRMHAS MKLFDSICNN KWFTDTSIIL FLNKKDLFEE KIKKSPLTIC YPEYAGSNTY EEAAAYIQCQ FED LNKRKD TKEIYTHFTC STDTKNVQFV FDAVTDVIIK NNLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Stromal cell-derived factor 1
| Macromolecule | Name: Stromal cell-derived factor 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.97846 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KPVSLSYRCP CRFFESHVAR ANVKHLKILN TPNCALQIVA RLKNNNRQVC IDPKLKWIQE YLEKALNK UniProtKB: Stromal cell-derived factor 1 |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.29 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 87963 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)