-検索条件
-検索結果
検索 (著者・登録者: jesse & im)の結果104件中、1から50件目までを表示しています

EMDB-50585: 
Icosahedral Encapsulin with a closed pore state

PDB-9fn9: 
Icosahedral Encapsulin with a closed pore state

EMDB-50586: 
CryoEM structure of Encapsulin::tdNfsB with an open pore state

PDB-9fna: 
CryoEM structure of Encapsulin::tdNfsB with an open pore state

EMDB-43250: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, global refinement

EMDB-43261: 
SARS-CoV-2 spike omicron (BA.1) ectodomain dimer-of-trimers in complex with SC27 Fab, global refinement

EMDB-44299: 
Cryo-EM structure of the desensitised ATP-bound human P2X1 receptor

EMDB-44370: 
Cryo-EM structure of the closed NF449-bound human P2X1 receptor

EMDB-43260: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement

EMDB-43315: 
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs

PDB-8vif: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement

PDB-8vke: 
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs

EMDB-16375: 
SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody

PDB-8c0y: 
SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody

EMDB-42970: 
Model and map from local refinement of a CAB-A17 - Omicron Ba.1 spike complex

PDB-8v4f: 
Model and map from local refinement of a CAB-A17 - Omicron Ba.1 spike complex

EMDB-16397: 
SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody

PDB-8c2r: 
SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)

PDB-7y71: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7

PDB-7y72: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175

PDB-7uhb: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)

PDB-7uhc: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
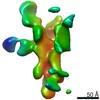
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
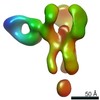
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
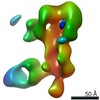
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
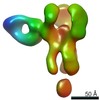
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA

EMDB-25646: 
Negative stain map of polyclonal Fab 241.14 binding the RBS epitope of H1 HA

EMDB-25655: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA

PDB-7t3d: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA

EMDB-24497: 
Yeast CTP Synthase (URA8) tetramer bound to ATP/UTP at neutral pH

EMDB-24512: 
Yeast CTP Synthase (URA8) Filament bound to ATP/UTP at low pH

EMDB-24516: 
Yeast CTP Synthase (URA8) Filament bound to CTP at low pH

EMDB-24560: 
Yeast CTP Synthase (Ura7) filament bound to CTP at low pH

EMDB-24566: 
Substrate-bound Ura7 filament at low pH
ページ:
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
