+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody / Spike / Complex / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Enterobacteria phage T4 (virus) / Enterobacteria phage T4 (virus) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
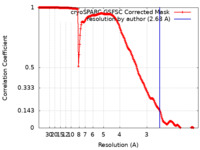
| Method | single particle reconstruction / cryo EM / Resolution: 2.68 Å | |||||||||
 Authors Authors | Das H / Hallberg BM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2024 Journal: Cell Rep Med / Year: 2024Title: Structural basis of broad SARS-CoV-2 cross-neutralization by affinity-matured public antibodies. Authors: Daniel J Sheward / Pradeepa Pushparaj / Hrishikesh Das / Allison J Greaney / Changil Kim / Sungyong Kim / Leo Hanke / Erik Hyllner / Robert Dyrdak / Jimin Lee / Xaquin Castro Dopico / Pia ...Authors: Daniel J Sheward / Pradeepa Pushparaj / Hrishikesh Das / Allison J Greaney / Changil Kim / Sungyong Kim / Leo Hanke / Erik Hyllner / Robert Dyrdak / Jimin Lee / Xaquin Castro Dopico / Pia Dosenovic / Thomas P Peacock / Gerald M McInerney / Jan Albert / Martin Corcoran / Jesse D Bloom / Ben Murrell / Gunilla B Karlsson Hedestam / B Martin Hällberg /      Abstract: Descendants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant now account for almost all SARS-CoV-2 infections. The Omicron variant and its sublineages have spike ...Descendants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant now account for almost all SARS-CoV-2 infections. The Omicron variant and its sublineages have spike glycoproteins that are highly diverged from the pandemic founder and first-generation vaccine strain, resulting in significant evasion from monoclonal antibody therapeutics and vaccines. Understanding how commonly elicited antibodies can broaden to cross-neutralize escape variants is crucial. We isolate IGHV3-53, using "public" monoclonal antibodies (mAbs) from an individual 7 months post infection with the ancestral virus and identify antibodies that exhibit potent and broad cross-neutralization, extending to the BA.1, BA.2, and BA.4/BA.5 sublineages of Omicron. Deep mutational scanning reveals these mAbs' high resistance to viral escape. Structural analysis via cryoelectron microscopy of a representative broadly neutralizing antibody, CAB-A17, in complex with the Omicron BA.1 spike highlights the structural underpinnings of this broad neutralization. By reintroducing somatic hypermutations into a germline-reverted CAB-A17, we delineate the role of affinity maturation in the development of cross-neutralization by a public class of antibodies. #1:  Journal: Biochem Biophys Res Commun / Year: 1975 Journal: Biochem Biophys Res Commun / Year: 1975Title: Delineation of the intimate details of the backbone conformation of pyridine nucleotide coenzymes in aqueous solution. Authors: Bose KS / Sarma RH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16375.map.gz emd_16375.map.gz | 412.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16375-v30.xml emd-16375-v30.xml emd-16375.xml emd-16375.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16375_fsc.xml emd_16375_fsc.xml | 19.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16375.png emd_16375.png | 15.6 KB | ||
| Filedesc metadata |  emd-16375.cif.gz emd-16375.cif.gz | 6.3 KB | ||
| Others |  emd_16375_half_map_1.map.gz emd_16375_half_map_1.map.gz emd_16375_half_map_2.map.gz emd_16375_half_map_2.map.gz | 764.7 MB 764.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16375 http://ftp.pdbj.org/pub/emdb/structures/EMD-16375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16375 | HTTPS FTP |
-Related structure data
| Related structure data |  8c0yMC  8c2rC  8v4fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16375.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16375.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16375_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16375_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV2 Omicron BA.1 Spike in complex with CAB-A17 antibody
| Entire | Name: SARS-CoV2 Omicron BA.1 Spike in complex with CAB-A17 antibody |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV2 Omicron BA.1 Spike in complex with CAB-A17 antibody
| Supramolecule | Name: SARS-CoV2 Omicron BA.1 Spike in complex with CAB-A17 antibody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 95.7 KDa |
-Supramolecule #2: SARS-CoV-2 Omicron RBD
| Supramolecule | Name: SARS-CoV-2 Omicron RBD / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: CAB-A17 Antibody (Fab)
| Supramolecule | Name: CAB-A17 Antibody (Fab) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
-Macromolecule #1: Spike protein S2'
| Macromolecule | Name: Spike protein S2' / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.942738 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAP FFTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGNIADYN YKLPDDFTGC VIAWNSNKLD SKVSGNYNYL YRLFRKSNLK PFERDISTEI YQAGNKPCNG VAGFNCYFPL R SYSFRPTY ...String: NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAP FFTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGNIADYN YKLPDDFTGC VIAWNSNKLD SKVSGNYNYL YRLFRKSNLK PFERDISTEI YQAGNKPCNG VAGFNCYFPL R SYSFRPTY GVGHQPYRVV VLSFELLHAP ATVCG UniProtKB: Spike glycoprotein |
-Macromolecule #2: CAB-A17 antibody (variable)
| Macromolecule | Name: CAB-A17 antibody (variable) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.395632 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAS LSCRASQSLS TYLAWYQQKP GQAPRLLIFG ASSRASGIPD RFSGGGSGTD FTLTISRLEP EDFAVYYCQ QYGSSPRTFG QGTKVEI |
-Macromolecule #3: CAB-A17 antibody (variable)
| Macromolecule | Name: CAB-A17 antibody (variable) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.839197 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVHLVESGGG LIQPGGSLRL SCAASEFIVS ANYMSWVRQA PGEGLQWVSV IYPGGSTFYA ESVKGRFTIS RDNSRNTLYL QMNSLRAED TGVYYCARDY GDFYFDYWGQ GTLVTVS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 25652 / Average exposure time: 1.5 sec. / Average electron dose: 54.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Calibrated defocus min: 0.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)