-Search query
-Search result
Showing 1 - 50 of 183 items for (author: holm & m)

EMDB-18202: 
Copper-transporting ATPase HMA4 in E1 state apo

EMDB-18203: 
Copper-transporting ATPase HMA4 in E1 state with Cu
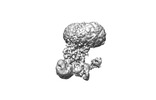
EMDB-18204: 
Copper-transporting ATPase HMA4 in E2P state with AlF

EMDB-18205: 
Copper-transporting ATPase HMA4 in E2P state with BeF

PDB-8q73: 
Copper-transporting ATPase HMA4 in E1 state apo

PDB-8q74: 
Copper-transporting ATPase HMA4 in E1 state with Cu

PDB-8q75: 
Copper-transporting ATPase HMA4 in E2P state with AlF

PDB-8q76: 
Copper-transporting ATPase HMA4 in E2P state with BeF

EMDB-41125: 
Zophobas morio black wasting virus strain NJ2-molitor virion structure

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
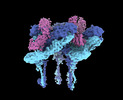
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

EMDB-17627: 
Recombinant Ena3A L-Type endospore appendages

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
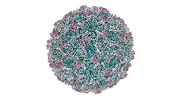
EMDB-41106: 
Zophobas morio black wasting virus strain UT-morio virion structure
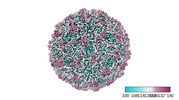
EMDB-41301: 
Zophobas morio black wasting virus strain OR-molitor virion structure

EMDB-40299: 
Consensus refinement of the h12-LOX in a dimeric form

EMDB-40300: 
The local refinement map of a "closed" subunit of a 12-LOX dimer

EMDB-40301: 
The local refinement map of an "open" subunit of a 12-LOX dimer

EMDB-40302: 
The consensus map of a 12-LOX hexamer

EMDB-40304: 
The local refinement map of a single subunit of a 12-LOX hexamer

EMDB-40039: 
The structure of h12-LOX in monomeric form

EMDB-40040: 
The structure of h12-LOX in dimeric form

EMDB-40041: 
The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid

EMDB-40042: 
The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA

PDB-8ghb: 
The structure of h12-LOX in monomeric form

PDB-8ghc: 
The structure of h12-LOX in dimeric form

PDB-8ghd: 
The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid

PDB-8ghe: 
The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA
EMDB-41108: 
Zophobas morio black wasting virus strain UT-morio empty capsid structure
EMDB-41131: 
Zophobas morio black wasting virus strain OR-molitor empty capsid structure
EMDB-28067: 
CryoEM reconstruction of tri-specific 298-52-80 Multabody (no symmetry applied)

EMDB-28068: 
CryoEM reconstruction of the tri-specific 298-52-80 Multabody (octahedral symmetry applied)

EMDB-29757: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state)

EMDB-29758: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state)

EMDB-29759: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state)

EMDB-29760: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state)

EMDB-29766: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (60S Focus refined map)

EMDB-29768: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (40S Focus refined map)

EMDB-29771: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2)

EMDB-29782: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (80S consensus refined structure)

EMDB-40205: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure)

PDB-8g5y: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state)

PDB-8g5z: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state)

PDB-8g60: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state)

PDB-8g61: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state)

PDB-8g6j: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2)

PDB-8glp: 
mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure)
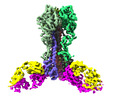
EMDB-27139: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
