[English] 日本語
 Yorodumi
Yorodumi- EMDB-40041: The structure of h12-LOX in hexameric form bound to inhibitor ML3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid | |||||||||
 Map data Map data | This is a composite map for the entire 12-LOX hexamer. It was created by applying D3 symmetry to a locally refined map of a monomeric subunit. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lipoxygenase / platelets / lipid-modifying enzyme / lipid oxidation / OXIDOREDUCTASE / OXIDOREDUCTASE-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationunsaturated fatty acid metabolic process / hepoxilin-epoxide hydrolase activity / leukotriene A4 metabolic process / lipoxin B4 biosynthetic process / Synthesis of Hepoxilins (HX) and Trioxilins (TrX) / Biosynthesis of DPAn-6 SPMs / Hydrolases; Acting on ether bonds; Ether hydrolases / arachidonate 12-lipoxygenase / lipoxin A4 biosynthetic process / Biosynthesis of DHA-derived SPMs ...unsaturated fatty acid metabolic process / hepoxilin-epoxide hydrolase activity / leukotriene A4 metabolic process / lipoxin B4 biosynthetic process / Synthesis of Hepoxilins (HX) and Trioxilins (TrX) / Biosynthesis of DPAn-6 SPMs / Hydrolases; Acting on ether bonds; Ether hydrolases / arachidonate 12-lipoxygenase / lipoxin A4 biosynthetic process / Biosynthesis of DHA-derived SPMs / Biosynthesis of DPAn-3-derived maresins / arachidonate 15-lipoxygenase / arachidonate 12(S)-lipoxygenase activity / arachidonate 15-lipoxygenase activity / Synthesis of 12-eicosatetraenoic acid derivatives / linoleate 13S-lipoxygenase activity / negative regulation of muscle cell apoptotic process / Biosynthesis of Lipoxins (LX) / lipoxygenase pathway / Oxidoreductases; Acting on single donors with incorporation of molecular oxygen (oxygenases); With incorporation of two atoms of oxygen / arachidonate metabolic process / lipid oxidation / hepoxilin biosynthetic process / linoleic acid metabolic process / negative regulation of platelet aggregation / superoxide anion generation / fatty acid oxidation / establishment of skin barrier / lipid metabolic process / sarcolemma / iron ion binding / extracellular exosome / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Black KA / Mobbs JI / Venugopal H / Thal DM / Glukhova A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Blood / Year: 2023 Journal: Blood / Year: 2023Title: Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors. Authors: Jesse I Mobbs / Katrina A Black / Michelle Tran / Wessel A C Burger / Hariprasad Venugopal / Theodore R Holman / Michael Holinstat / David M Thal / Alisa Glukhova /   Abstract: Human 12-lipoxygenase (12-LOX) is a key enzyme involved in platelet activation, and the regulation of its activity has been targeted for the treatment of heparin-induced thrombocytopenia. Despite the ...Human 12-lipoxygenase (12-LOX) is a key enzyme involved in platelet activation, and the regulation of its activity has been targeted for the treatment of heparin-induced thrombocytopenia. Despite the clinical importance of 12-LOX, the exact mechanisms by which it affects platelet activation are not fully understood, and the lack of structural information has limited drug discovery efforts. In this study, we used single-particle cryo-electron microscopy to determine high-resolution structures (1.7-2.8 Å) of human 12-LOX. Our results showed that 12-LOX can exist in multiple oligomeric states, from monomer to hexamer, which may affect its catalytic activity and membrane association. We also identified different conformations within the 12-LOX dimer, which likely represent different time points in its catalytic cycle. Furthermore, we identified small molecules bound to 12-LOX. The active site of the 12-LOX tetramer was occupied by an endogenous 12-LOX inhibitor, a long-chain acyl coenzyme A. In addition, we found that the 12-LOX hexamer can simultaneously bind to arachidonic acid and ML355, a selective 12-LOX inhibitor that has passed a phase 1 clinical trial for the treatment of heparin-induced thrombocytopenia and received a fast-track designation by the Food and Drug Administration. Overall, our findings provide novel insights into the assembly of 12-LOX oligomers, their catalytic mechanism, and small molecule binding, paving the way for further drug development targeting the 12-LOX enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40041.map.gz emd_40041.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40041-v30.xml emd-40041-v30.xml emd-40041.xml emd-40041.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40041.png emd_40041.png | 192.6 KB | ||
| Filedesc metadata |  emd-40041.cif.gz emd-40041.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40041 http://ftp.pdbj.org/pub/emdb/structures/EMD-40041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40041 | HTTPS FTP |
-Related structure data
| Related structure data |  8ghdMC  8ghbC  8ghcC  8gheC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40041.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40041.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a composite map for the entire 12-LOX hexamer. It was created by applying D3 symmetry to a locally refined map of a monomeric subunit. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
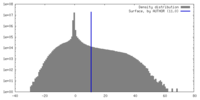
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Hexameric human 12-Lipoxygenase in complex with inhibitor ML355 a...
| Entire | Name: Hexameric human 12-Lipoxygenase in complex with inhibitor ML355 and arachidonic acid |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric human 12-Lipoxygenase in complex with inhibitor ML355 a...
| Supramolecule | Name: Hexameric human 12-Lipoxygenase in complex with inhibitor ML355 and arachidonic acid type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Polyunsaturated fatty acid lipoxygenase ALOX12
| Macromolecule | Name: Polyunsaturated fatty acid lipoxygenase ALOX12 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: Oxidoreductases; Acting on single donors with incorporation of molecular oxygen (oxygenases); With incorporation of two atoms of oxygen |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.582703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHGRY RIRVATGAWL FSGSYNRVQL WLVGTRGEAE LELQLRPARG EEEEFDHDVA EDLGLLQFVR LRKHHWLVDD AWFCDRITV QGPGACAEVA FPCYRWVQGE DILSLPEGTA RLPGDNALDM FQKHREKELK DRQQIYCWAT WKEGLPLTIA A DRKDDLPP ...String: MHHHHHHGRY RIRVATGAWL FSGSYNRVQL WLVGTRGEAE LELQLRPARG EEEEFDHDVA EDLGLLQFVR LRKHHWLVDD AWFCDRITV QGPGACAEVA FPCYRWVQGE DILSLPEGTA RLPGDNALDM FQKHREKELK DRQQIYCWAT WKEGLPLTIA A DRKDDLPP NMRFHEEKRL DFEWTLKAGA LEMALKRVYT LLSSWNCLED FDQIFWGQKS ALAEKVRQCW QDDELFSYQF LN GANPMLL RRSTSLPSRL VLPSGMEELQ AQLEKELQNG SLFEADFILL DGIPANVIRG EKQYLAAPLV MLKMEPNGKL QPM VIQIQP PSPSSPTPTL FLPSDPPLAW LLAKSWVRNS DFQLHEIQYH LLNTHLVAEV IAVATMRCLP GLHPIFKFLI PHIR YTMEI NTRARTQLIS DGGIFDKAVS TGGGGHVQLL RRAAAQLTYC SLCPPDDLAD RGLLGLPGAL YAHDALRLWE IIARY VEGI VHLFYQRDDI VKGDPELQAW CREITEVGLC QAQDRGFPVS FQSQSQLCHF LTMCVFTCTA QHAAINQGQL DWYAWV PNA PCTMRMPPPT TKEDVTMATV MGSLPDVRQA CLQMAISWHL SRRQPDMVPL GHHKEKYFSG PKPKAVLNQF RTDLEKL EK EITARNEQLD WPYEYLKPSC IENSVTI UniProtKB: Polyunsaturated fatty acid lipoxygenase ALOX12 |
-Macromolecule #2: FE (II) ION
| Macromolecule | Name: FE (II) ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: FE2 |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #3: N-(1,3-benzothiazol-2-yl)-4-{[(2-hydroxy-3-methoxyphenyl)methyl]a...
| Macromolecule | Name: N-(1,3-benzothiazol-2-yl)-4-{[(2-hydroxy-3-methoxyphenyl)methyl]amino}benzene-1-sulfonamide type: ligand / ID: 3 / Number of copies: 6 / Formula: ZR5 |
|---|---|
| Molecular weight | Theoretical: 441.523 Da |
| Chemical component information |  ChemComp-ZR5: |
-Macromolecule #4: ARACHIDONIC ACID
| Macromolecule | Name: ARACHIDONIC ACID / type: ligand / ID: 4 / Number of copies: 6 / Formula: ACD |
|---|---|
| Molecular weight | Theoretical: 304.467 Da |
| Chemical component information | 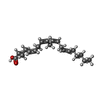 ChemComp-ACD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: FLEXIBLE FIT |
| Output model |  PDB-8ghd: |
-Atomic model buiding 2
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8ghd: |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)