-Search query
-Search result
Showing 1 - 50 of 680 items for (author: ho & cm)

EMDB-43139: 
SARS-CoV-2 Spike S2 bound to Fab 54043-5

EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)

EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

EMDB-43762: 
Aca2 from Pectobacterium phage ZF40 bound to RNA

EMDB-43097: 
Simulation-driven design of prefusion stabilized SARS-CoV-2 spike S2 antigen
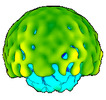
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry
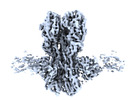
EMDB-41874: 
CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06

EMDB-50356: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-50357: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry
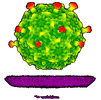
EMDB-50359: 
Rhodobacter microvirus Ebor attached to B10 host cell reconstructed by single particle analysis with applied C5 symmetry
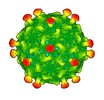
EMDB-50360: 
Rhodobacter microvirus Ebor attached to the outer membrane vesicle

EMDB-50361: 
Rhodobacter microvirus Ebor attached to the host cell reconstructed by subtomogram averaging

EMDB-41248: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

EMDB-41249: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

PDB-8th3: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

PDB-8th4: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

EMDB-19426: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137

EMDB-19427: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I

EMDB-19428: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II

EMDB-19429: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III

EMDB-19406: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12

EMDB-19407: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24

PDB-8rox: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12

PDB-8roy: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24

EMDB-41277: 
Cryo-EM structure of a SUR1/Kir6.2-Q52R ATP-sensitive potassium channel in the presence of PIP2 in the open conformation

EMDB-41278: 
Cryo-EM structure of a SUR1/Kir6.2-Q52R ATP-sensitive potassium channel in the presence of PIP2 in the open conformation

EMDB-43766: 
Kir6.2-Q52R/SUR1 apo closed channel

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane
EMDB-19054: 
Escherichia coli paused disome complex (Non-rotated disome interface class 1)

EMDB-17274: 
Escherichia coli paused disome complex (decoding | rotated PRE)

EMDB-17275: 
Escherichia coli paused disome complex (decoding | closed non-rot. PRE)

EMDB-17276: 
Escherichia coli paused disome complex (decoding | POST)

EMDB-17277: 
Escherichia coli paused disome complex (partially accommodated A-tRNA | closed non-rotated PRE)

EMDB-17278: 
Escherichia coli paused disome complex (partially accommodated A-tRNA | rotated PRE)

EMDB-17279: 
Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)

EMDB-17280: 
Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)

EMDB-17281: 
Escherichia coli paused disome complex (open non-rotated PRE | rotated PRE)

EMDB-17282: 
Escherichia coli paused disome complex (open non-rotated PRE | POST)

EMDB-17283: 
Escherichia coli paused disome complex (closed non-rotated PRE | closed non-rotated PRE)

EMDB-17284: 
Escherichia coli paused disome complex (closed non-rotated PRE | rotated PRE)

EMDB-17285: 
Escherichia coli paused disome complex (closed non-rotated PRE | POST)

EMDB-17286: 
Escherichia coli paused disome complex (rotated-PRE-1 | closed non-rotated PRE)

EMDB-17288: 
Escherichia coli paused disome complex (rotated-PRE-1 | rotated PRE)

EMDB-17289: 
Cryo-EM reconstruction of the E. coli disome complex (rotated-PRE-1 | POST)

EMDB-17291: 
Escherichia coli paused disome complex (rotated-PRE-2 | closed non-rotated PRE)

EMDB-17431: 
Escherichia coli paused disome complex (rotated-PRE-2 | rotated PRE)

EMDB-17432: 
Escherichia coli paused disome complex (rotated-PRE-2 | POST)

EMDB-17433: 
Escherichia coli paused disome complex (POST | closed non-rotated PRE)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
