-Search query
-Search result
Showing 1 - 50 of 720 items for (author: he & bb)
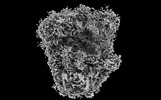
EMDB-41485: 
Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
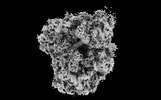
EMDB-41486: 
Subtomogram averaged decoding-1 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41487: 
Subtomogram averaged decoding-2 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41488: 
Subtomogram averaged classical iPRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
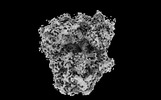
EMDB-41489: 
Subtomogram averaged rotated-1 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41490: 
Subtomogram averaged rotated-2 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41491: 
Subtomogram averaged translocation state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
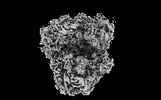
EMDB-41492: 
Subtomogram averaged POST state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41493: 
Subtomogram averaged unloaded state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
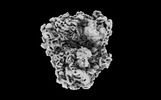
EMDB-41494: 
Subtomogram averaged non-rotated 80S ribosome of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
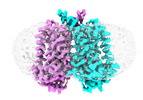
EMDB-43712: 
Human EBP complexed with compound 1
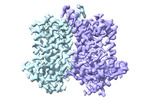
EMDB-43713: 
Human EBP complexed with compound 3a

PDB-8w0r: 
Human EBP complexed with compound 1

PDB-8w0s: 
Human EBP complexed with compound 3a

EMDB-50019: 
cryoEM structure of Photosystem II averaged across S2-S3 states at 1.71 Angstrom resolution

PDB-9evx: 
cryoEM structure of Photosystem II averaged across S2-S3 states at 1.71 Angstrom resolution
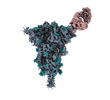
EMDB-40812: 
Structure of SARS-CoV-2 (HP-GSAS-Mut7) spike in complex with TXG-0078 Fab -Conformation 1

EMDB-40813: 
Structure of SARS-CoV-2 (HP-GSAS-Mut7) spike in complex with TXG-0078 Fab -Conformation 2

EMDB-36646: 
Cryo-EM structure of GeoCas9-sgRNA-dsDNA ternary complex

EMDB-36650: 
Cryo-EM structure of GeoCas9-sgRNA binary complex

EMDB-40261: 
DDB1/CRBN in complex with ARV-471 and the ER ligand-binding domain

EMDB-44642: 
Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex

EMDB-44643: 
Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex

PDB-9bkj: 
Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex

PDB-9bkk: 
Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex

EMDB-40814: 
Local refinement of SARS-CoV-2 (HP-GSAS-Mut7) spike NTD in complex with TXG-0078 Fab

PDB-8swh: 
Local refinement of SARS-CoV-2 (HP-GSAS-Mut7) spike NTD in complex with TXG-0078 Fab

EMDB-43753: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain

PDB-8w2o: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain
EMDB-42464: 
chEnv TTT protein in complex with 43A2 Fab

EMDB-42468: 
chEnv TTT protein in complex with CM01A Fab

EMDB-17924: 
Cryo-EM structure of human Elp123 in complex with tRNA, acetyl-CoA, 5'-deoxyadenosine and methionine

EMDB-17925: 
Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosine and methionine

EMDB-17926: 
Cryo-EM structure of human Elp123 in complex with tRNA, S-ethyl-CoA, 5'-deoxyadenosine and methionine

EMDB-17927: 
Cryo-EM structure of human Elp123 in complex with tRNA, desulpho-CoA, 5'-deoxyadenosine and methionine

PDB-8ptx: 
Cryo-EM structure of human Elp123 in complex with tRNA, acetyl-CoA, 5'-deoxyadenosine and methionine

PDB-8pty: 
Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosine and methionine

PDB-8ptz: 
Cryo-EM structure of human Elp123 in complex with tRNA, S-ethyl-CoA, 5'-deoxyadenosine and methionine

PDB-8pu0: 
Cryo-EM structure of human Elp123 in complex with tRNA, desulpho-CoA, 5'-deoxyadenosine and methionine

EMDB-43664: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines
EMDB-43665: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (cH125 TTT)

EMDB-43666: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (H2/1 GCN4)

EMDB-43668: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (H5/1 GCN4)

EMDB-43669: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines. H5 GCN4

EMDB-16229: 
Cryo-EM structure of the bacterial replication origin opening basal unwinding system

PDB-8btg: 
Cryo-EM structure of the bacterial replication origin opening basal unwinding system

EMDB-40984: 
5TU-t1 - heterodimeric triplet polymerase ribozyme

PDB-8t2p: 
5TU-t1 - heterodimeric triplet polymerase ribozyme

EMDB-43271: 
Cryo-EM structure of the Helicobacter pylori VacA hexamer that was detergent solubilized from membrane, C6 symmetry applied

EMDB-43272: 
Cryo-EM structure of Helicobacter pylori VacA hexamer that was detergent solubilized form membrane, no symmetry applied
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
