[English] 日本語
 Yorodumi
Yorodumi- EMDB-43666: Triple tandem trimer immunogens for HIV-1 and influenza nucleic a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (H2/1 GCN4) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hemagglutinin / CR9114 Fab / VIRUS | |||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 19.52 Å | |||||||||
 Authors Authors | Ferguson JA / Leon AN / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2024 Journal: NPJ Vaccines / Year: 2024Title: Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines. Authors: Iván Del Moral-Sánchez / Edmund G Wee / Yuejiao Xian / Wen-Hsin Lee / Joel D Allen / Alba Torrents de la Peña / Rebeca Fróes Rocha / James Ferguson / André N León / Sylvie Koekkoek / ...Authors: Iván Del Moral-Sánchez / Edmund G Wee / Yuejiao Xian / Wen-Hsin Lee / Joel D Allen / Alba Torrents de la Peña / Rebeca Fróes Rocha / James Ferguson / André N León / Sylvie Koekkoek / Edith E Schermer / Judith A Burger / Sanjeev Kumar / Robby Zwolsman / Mitch Brinkkemper / Aafke Aartse / Dirk Eggink / Julianna Han / Meng Yuan / Max Crispin / Gabriel Ozorowski / Andrew B Ward / Ian A Wilson / Tomáš Hanke / Kwinten Sliepen / Rogier W Sanders /      Abstract: Recombinant native-like HIV-1 envelope glycoprotein (Env) trimers are used in candidate vaccines aimed at inducing broadly neutralizing antibodies. While state-of-the-art SOSIP or single-chain Env ...Recombinant native-like HIV-1 envelope glycoprotein (Env) trimers are used in candidate vaccines aimed at inducing broadly neutralizing antibodies. While state-of-the-art SOSIP or single-chain Env designs can be expressed as native-like trimers, undesired monomers, dimers and malformed trimers that elicit non-neutralizing antibodies are also formed, implying that these designs could benefit from further modifications for gene-based vaccination approaches. Here, we describe the triple tandem trimer (TTT) design, in which three Env protomers are genetically linked in a single open reading frame and express as native-like trimers. Viral vectored Env TTT induced similar neutralization titers but with a higher proportion of trimer-specific responses. The TTT design was also applied to generate influenza hemagglutinin (HA) trimers without the need for trimerization domains. Additionally, we used TTT to generate well-folded chimeric Env and HA trimers that harbor protomers from three different strains. In summary, the TTT design is a useful platform for the design of HIV-1 Env and influenza HA immunogens for a multitude of vaccination strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43666.map.gz emd_43666.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43666-v30.xml emd-43666-v30.xml emd-43666.xml emd-43666.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43666.png emd_43666.png | 39.4 KB | ||
| Filedesc metadata |  emd-43666.cif.gz emd-43666.cif.gz | 4.3 KB | ||
| Others |  emd_43666_half_map_1.map.gz emd_43666_half_map_1.map.gz emd_43666_half_map_2.map.gz emd_43666_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43666 http://ftp.pdbj.org/pub/emdb/structures/EMD-43666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43666 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43666.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43666.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43666_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
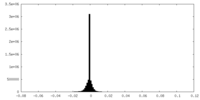
| Density Histograms |
-Half map: #2
| File | emd_43666_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
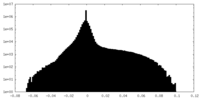
| Density Histograms |
- Sample components
Sample components
-Entire : Chimeric hemagglutinin in complex with CR9114 Fab.
| Entire | Name: Chimeric hemagglutinin in complex with CR9114 Fab. |
|---|---|
| Components |
|
-Supramolecule #1: Chimeric hemagglutinin in complex with CR9114 Fab.
| Supramolecule | Name: Chimeric hemagglutinin in complex with CR9114 Fab. / type: complex / ID: 1 / Parent: 0 / Details: (H2/1 GCN4) |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: Tris-Buffered Saline |
| Staining | Type: NEGATIVE / Material: Uranyl formate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: PDB2MRC PDBID 4m4y Low pass filtered to 20A |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 19.52 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 22735 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)