-Search query
-Search result
Showing 1 - 50 of 104 items for (author: hafenstein & sh)
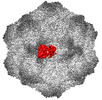
EMDB-26786: 
CPV Affinity Purified Polyclonal Fab A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
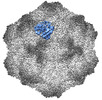
EMDB-26787: 
CPV Affinity Purified Polyclonal Fab B Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
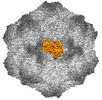
EMDB-26788: 
CPV Total-Fab Polyclonal A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
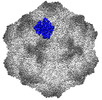
EMDB-26789: 
CPV Total-Fab Polyclonal B Site Fab (1 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
EMDB-26790: 
CPV Total-Fab Polyclonal B Site Fab (2 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

PDB-7utp: 
CPV Affinity Purified Polyclonal Fab A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

PDB-7utr: 
CPV Affinity Purified Polyclonal Fab B Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

PDB-7uts: 
CPV Total-Fab Polyclonal A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

PDB-7utu: 
CPV Total-Fab Polyclonal B Site Fab (1 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

PDB-7utv: 
CPV Total-Fab Polyclonal B Site Fab (2 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

EMDB-27943: 
9H2 Fab-poliovirus 1 complex
Method: single particle / : Charnesky AJ
EMDB-27947: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ

EMDB-27948: 
9H2 Fab-poliovirus 2 complex
Method: single particle / : Charnesky AJ
EMDB-27949: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ
EMDB-27950: 
9H2 Fab-Sabin poliovirus 2 complex
Method: single particle / : Charnesky AJ
EMDB-27951: 
9H2 Fab-Sabin poliovirus 1 complex
Method: single particle / : Charnesky AJ

EMDB-24741: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

PDB-7ryj: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

EMDB-23656: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23657: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23658: 
Canine parvovirus and Fab14 asymmetric reconstruction
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23659: 
Canine parvovirus asymmetric map
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

PDB-7m3l: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

PDB-7m3m: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

PDB-7m3n: 
Canine parvovirus and Fab14 asymmetric reconstruction
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

PDB-7m3o: 
Canine parvovirus asymmetric map
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-22640: 
Murine polyomavirus pentavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22641: 
Murine polyomavirus hexavalent capsomer with 8A7H5 Fab, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22642: 
Murine polyomavirus pentavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22643: 
Murine polyomavirus hexavalent capsomer, subparticle reconstruction
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22645: 
Murine polyomavirus (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22646: 
Murine polyomavirus with 8A7H5 Fab (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-20001: 
Structure of canine parvovirus in complex with transferrin receptor type-1
Method: single particle / : Lee H, Hafenstein S

EMDB-20002: 
Structure of canine parvovirus in complex with transferrin receptor type-1
Method: single particle / : Lee H, Hafenstein S

EMDB-20003: 
Transferrin-transferrin receptor complex bound to canine parvovirus capsid
Method: single particle / : Lee H, Hafenstein S

PDB-6oas: 
Structure of canine parvovirus in complex with transferrin receptor type-1
Method: single particle / : Lee H, Hafenstein S

EMDB-9363: 
Structure of M. spretus Endogenous Virus Element (EVE) Virus-like particle (VLP)
Method: single particle / : Callaway HM, Subramanian S

PDB-6nf9: 
Structure of M. spretus Endogenous Virus Element (EVE) Virus-like particle (VLP)
Method: single particle / : Callaway HM, Subramanian S

EMDB-7136: 
High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christenson ND, Hafenstein S

PDB-6bsp: 
High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christenson ND, Hafenstein S

PDB-6bt3: 
High-Resolution Structure Analysis of Antibody V5 Conformational Epitope on Human Papillomavirus 16
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-8243: 
Near-atomic resolution cryo-EM reconstruction of HPV16 complexed with Fab V5
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-6619: 
Electron cryo-microscopy of Human Papillomavirus
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-6620: 
Electron cryo-microscopy of Human Papillomavirus
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

PDB-5kep: 
High resolution cryo-EM maps of Human Papillomavirus 16 reveal L2 location and heparin-induced conformational changes
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

PDB-5keq: 
High resolution cryo-EM maps of Human papillomavirus 16 reveal L2 location and heparin-induced conformational changes
Method: single particle / : Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S

EMDB-8463: 
Deformed Wing Virus Assembly Intermediate Procapsid
Method: single particle / : Organtini LJ, Hafenstein S, Conway JF, Makhov AM, Dryden KA, Pizzorno MC

EMDB-8464: 
Deformed Wing Virus 80-S Like Particle
Method: single particle / : Organtini LJ, Hafenstein S, Conway JF, Makhov AM, Dryden KA, Pizzorno MC

EMDB-8371: 
Three-dimensional reconstruction of bat adenovirus C isolate 250-A
Method: single particle / : Hackenbrack NM, Hafenstein SL

EMDB-6636: 
The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs
Method: single particle / : Shingler KL, Lee H, Organtini LJ, Ashley RE, Subramanian S, Makhov AM, Conway JF, Hafenstein S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
