[English] 日本語
 Yorodumi
Yorodumi- EMDB-22643: Murine polyomavirus hexavalent capsomer, subparticle reconstruction -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22643 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Murine polyomavirus hexavalent capsomer, subparticle reconstruction | |||||||||||||||||||||
 Map data Map data | Murine polyomavirus hexavalent capsomer, subparticle reconstruction | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | polyomavirus / capsomer / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcaveolin-mediated endocytosis of virus by host cell / T=7 icosahedral viral capsid / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity Similarity search - Function | |||||||||||||||||||||
| Biological species |  Mus musculus polyomavirus 1 / Mus musculus polyomavirus 1 /  Murine polyomavirus strain A2 Murine polyomavirus strain A2 | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||
 Authors Authors | Goetschius DJ / Hafenstein SL | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Antibody escape by polyomavirus capsid mutation facilitates neurovirulence. Authors: Matthew D Lauver / Daniel J Goetschius / Colleen S Netherby-Winslow / Katelyn N Ayers / Ge Jin / Daniel G Haas / Elizabeth L Frost / Sung Hyun Cho / Carol M Bator / Stephanie M Bywaters / ...Authors: Matthew D Lauver / Daniel J Goetschius / Colleen S Netherby-Winslow / Katelyn N Ayers / Ge Jin / Daniel G Haas / Elizabeth L Frost / Sung Hyun Cho / Carol M Bator / Stephanie M Bywaters / Neil D Christensen / Susan L Hafenstein / Aron E Lukacher /  Abstract: JCPyV polyomavirus, a member of the human virome, causes progressive multifocal leukoencephalopathy (PML), an oft-fatal demyelinating brain disease in individuals receiving immunomodulatory therapies. ...JCPyV polyomavirus, a member of the human virome, causes progressive multifocal leukoencephalopathy (PML), an oft-fatal demyelinating brain disease in individuals receiving immunomodulatory therapies. Mutations in the major viral capsid protein, VP1, are common in JCPyV from PML patients (JCPyV-PML) but whether they confer neurovirulence or escape from virus-neutralizing antibody (nAb) in vivo is unknown. A mouse polyomavirus (MuPyV) with a sequence-equivalent JCPyV-PML VP1 mutation replicated poorly in the kidney, a major reservoir for JCPyV persistence, but retained the CNS infectivity, cell tropism, and neuropathology of the parental virus. This mutation rendered MuPyV resistant to a monoclonal Ab (mAb), whose specificity overlapped the endogenous anti-VP1 response. Using cryo-EM and a custom sub-particle refinement approach, we resolved an MuPyV:Fab complex map to 3.2 Å resolution. The structure revealed the mechanism of mAb evasion. Our findings demonstrate convergence between nAb evasion and CNS neurovirulence in vivo by a frequent JCPyV-PML VP1 mutation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22643.map.gz emd_22643.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22643-v30.xml emd-22643-v30.xml emd-22643.xml emd-22643.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22643_fsc.xml emd_22643_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22643.png emd_22643.png | 246.6 KB | ||
| Filedesc metadata |  emd-22643.cif.gz emd-22643.cif.gz | 6.1 KB | ||
| Others |  emd_22643_half_map_1.map.gz emd_22643_half_map_1.map.gz emd_22643_half_map_2.map.gz emd_22643_half_map_2.map.gz | 79.8 MB 79.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22643 http://ftp.pdbj.org/pub/emdb/structures/EMD-22643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22643 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22643 | HTTPS FTP |
-Related structure data
| Related structure data |  7k25MC  7k22C  7k23C  7k24C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22643.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22643.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Murine polyomavirus hexavalent capsomer, subparticle reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Murine polyomavirus hexavalent capsomer, subparticle reconstruction
| File | emd_22643_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Murine polyomavirus hexavalent capsomer, subparticle reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
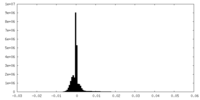
| Density Histograms |
-Half map: Murine polyomavirus hexavalent capsomer, subparticle reconstruction
| File | emd_22643_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Murine polyomavirus hexavalent capsomer, subparticle reconstruction | ||||||||||||
| Projections & Slices |
| ||||||||||||
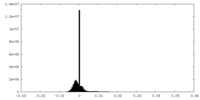
| Density Histograms |
- Sample components
Sample components
-Entire : Murine polyomavirus strain A2
| Entire | Name:  Murine polyomavirus strain A2 Murine polyomavirus strain A2 |
|---|---|
| Components |
|
-Supramolecule #1: Murine polyomavirus strain A2
| Supramolecule | Name: Murine polyomavirus strain A2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10636 / Sci species name: Murine polyomavirus strain A2 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Diameter: 450.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mus musculus polyomavirus 1 Mus musculus polyomavirus 1 |
| Molecular weight | Theoretical: 42.493172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APKRKSGVSK CETKCTKACP RPAPVPKLLI KGGMEVLDLV TGPDSVTEIE AFLNPRMGQP PTPESLTEGG QYYGWSRGIN LATSDTEDS PENNTLPTWS MAKLQLPMLN EDLTCDTLQM WEAVSVKTEV VGSGSLLDVH GFNKPTDTVN TKGISTPVEG S QYHVFAVG ...String: APKRKSGVSK CETKCTKACP RPAPVPKLLI KGGMEVLDLV TGPDSVTEIE AFLNPRMGQP PTPESLTEGG QYYGWSRGIN LATSDTEDS PENNTLPTWS MAKLQLPMLN EDLTCDTLQM WEAVSVKTEV VGSGSLLDVH GFNKPTDTVN TKGISTPVEG S QYHVFAVG GEPLDLQGLV TDARTKYKEE GVVTIKTITK KDMVNKDQVL NPISKAKLDK DGMYPVEIWH PDPAKNENTR YF GNYTGGT TTPPVLQFTN TLTTVLLDEN GVGPLCKGEG LYLSCVDIMG WRVTRNYDVH HWRGLPRYFK ITLRKRWVKN PYP MASLIS SLFNNMLPQV QGQPMEGENT QVEEVRVYDG TEPVPGDPDM TRYVDRFGKT KTVFPGN UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
Details: 10 mM HEPES pH 7.9, 1 mM CaCl2, 1 mM MgCl2, 5 mM KCl | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % | ||||||||||
| Details | MuPyV (2.8 mg/mL) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Homology model for Fab was generated using SwissModel. Initial models were docked into density in Chimera. Fab CDR loops were manually rebuilt in Coot. Iterative rounds of real space refinements (PHENIX) and manual adjustment (coot) were conducted to improve fit to density. | ||||||
| Refinement | Space: REAL | ||||||
| Output model |  PDB-7k25: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)