-検索条件
-検索結果
検索 (著者・登録者: gonzalez & jm)の結果全42件を表示しています

EMDB-42495: 
E. coli 70S ribosome with unmodified tRNAPro(GGG) bound to slippery P-site CCC-C codon and tRNAVal(UAC) in the A site

EMDB-42541: 
E. coli 70S ribosome with unmodified lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the 0 frame

EMDB-42714: 
E. coli 70S ribosome with unmodified Lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the +1 mRNA reading frame

EMDB-42721: 
E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon

EMDB-42832: 
E. coli 70S ribosome with unmodified e*/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon

EMDB-42840: 
E. coli 70S ribosome with unmodified e*/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon

EMDB-42852: 
E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon

EMDB-43306: 
cryo-EM structure of a non-neutralizing antibody V3-21 in complex with SARS-CoV-2 spike

EMDB-43314: 
cryo-EM structure of a non-neutralizing antibody V6-12 in complex with SARS-CoV-2 spike

EMDB-43255: 
Protective effect of human non-neutralizing cross-reactive spike antibodies elicited by SARS-CoV-2 mRNA vaccination

EMDB-14250: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment

EMDB-14271: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment (local refinement)

EMDB-13233: 
Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2

EMDB-4887: 
Cryo-EM reconstruction of Thermus thermophilus bactofilin double helical filaments

EMDB-3619: 
RnQV1-W1118 empty capsid

EMDB-3437: 
Three-dimensional reconstruction of W1075 strain Rosellinia necatrix quadrivirus 1

EMDB-3438: 
Three-dimensional reconstruction of W1118 strain Rosellinia necatrix quadrivirus 1

EMDB-3753: 
Negative stain 3D single particle reconstruction of the isolated surface adhesin P140/P110 complex of Mycoplasma genitalium.

EMDB-3756: 
Sub-tomogram average of the surface adhesin (NAP) complex from Mycoplasma genitalium cells by cryo-electron tomography.

EMDB-2573: 
SA11 Rotavirus non-trypsinized Triple Layered Particle

EMDB-2574: 
SA11 Rotavirus trypsinized Triple Layered Particle

EMDB-2575: 
SA11 Rotavirus non-trypsinized Triple Layered Particle grown in absence of leupetin

EMDB-2576: 
OSU Rotavirus non-trypsinized Triple Layered Particle

EMDB-2577: 
OSU Rotavirus trypsinized Triple Layered Particle
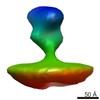
EMDB-2578: 
Class 1 averaged spike from SA11 Rotavirus non-trypsinized Triple Layered Particle

EMDB-2579: 
Class 2 averaged spike from SA11 Rotavirus non-trypsinized Triple Layered Particle
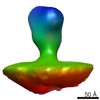
EMDB-2580: 
Class 1 averaged spike from SA11 Rotavirus trypsinized Triple Layered Particle

EMDB-5600: 
Penicillium Chrysogenum Virus (PcV) capsid structure

EMDB-5713: 
Electron microscopy of negatively-stained gp12 tubular protein of T7 bacteriophage

EMDB-5689: 
Cryo-electron microscopy of T7 tail complex

EMDB-5690: 
Cryo-electron microscopy of T7 tail complex formed by gp8, gp11, and gp12 proteins

EMDB-2062: 
Cryphonectria nitschkei Virus (CnV)capsid

EMDB-1933: 
wt Rabbit Hemorrhagic Disease Virus (RHDV)capsid

EMDB-1934: 
L17 Rabbit Hemorrhagic Disease Virus (RHDV)capsid

EMDB-1935: 
L42 Rabbit Hemorrhagic Disease Virus (RHDV)capsid

EMDB-1936: 
N42 Rabbit Hemorrhagic Disease Virus (RHDV) T3 capsid

EMDB-1937: 
N42 Rabbit Hemorrhagic Disease Virus (RHDV) T4 capsid

EMDB-1938: 
C42 Rabbit Hemorrhagic Disease Virus (RHDV) capsid

EMDB-1939: 
Rabbit Hemorrhagic Disease Virus (RHDV)capsid calculated from merged wt, L17 and N42 data sets

EMDB-1610: 
7.5 Amstrong resolution cryo-electron microscopy reconstruction of Penicillium chrysogenum virus (PcV)

EMDB-1611: 
7.5 Amstrong resolution cryo-electron microscopy reconstruction of Penicillium chrysogenum virus (PcV)

EMDB-1231: 
Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery.
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
