+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3619 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RnQV1-W1118 empty capsid | |||||||||
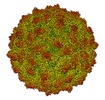
 Map data Map data | Rosellinia necatrix quadrivirus 1 strain W1118 (RnQV1-1118), empty particle | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Capsid protein / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Rosellinia necatrix quadrivirus 1 Rosellinia necatrix quadrivirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Mata CP / Luque D / Gomez-Blanco J / Rodriguez JM / Suzuki N / Ghabrial SA / Carrascosa JL / Trus BL / Caston JR | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2017 Journal: PLoS Pathog / Year: 2017Title: Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses. Authors: Carlos P Mata / Daniel Luque / Josué Gómez-Blanco / Javier M Rodríguez / José M González / Nobuhiro Suzuki / Said A Ghabrial / José L Carrascosa / Benes L Trus / José R Castón /    Abstract: Unlike their counterparts in bacterial and higher eukaryotic hosts, most fungal viruses are transmitted intracellularly and lack an extracellular phase. Here we determined the cryo-EM structure at 3. ...Unlike their counterparts in bacterial and higher eukaryotic hosts, most fungal viruses are transmitted intracellularly and lack an extracellular phase. Here we determined the cryo-EM structure at 3.7 Å resolution of Rosellinia necatrix quadrivirus 1 (RnQV1), a fungal double-stranded (ds)RNA virus. RnQV1, the type species of the family Quadriviridae, has a multipartite genome consisting of four monocistronic segments. Whereas most dsRNA virus capsids are based on dimers of a single protein, the ~450-Å-diameter, T = 1 RnQV1 capsid is built of P2 and P4 protein heterodimers, each with more than 1000 residues. Despite a lack of sequence similarity between the two proteins, they have a similar α-helical domain, the structural signature shared with the lineage of the dsRNA bluetongue virus-like viruses. Domain insertions in P2 and P4 preferential sites provide additional functions at the capsid outer surface, probably related to enzyme activity. The P2 insertion has a fold similar to that of gelsolin and profilin, two actin-binding proteins with a function in cytoskeleton metabolism, whereas the P4 insertion suggests protease activity involved in cleavage of the P2 383-residue C-terminal region, absent in the mature viral particle. Our results indicate that the intimate virus-fungus partnership has altered the capsid genome-protective and/or receptor-binding functions. Fungal virus evolution has tended to allocate enzyme activities to the virus capsid outer surface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3619.map.gz emd_3619.map.gz | 209.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3619-v30.xml emd-3619-v30.xml emd-3619.xml emd-3619.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3619.png emd_3619.png | 277.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3619 http://ftp.pdbj.org/pub/emdb/structures/EMD-3619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3619 | HTTPS FTP |
-Related structure data
| Related structure data |  5nd1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3619.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3619.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Rosellinia necatrix quadrivirus 1 strain W1118 (RnQV1-1118), empty particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rosellinia necatrix quadrivirus 1
| Entire | Name:  Rosellinia necatrix quadrivirus 1 Rosellinia necatrix quadrivirus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Rosellinia necatrix quadrivirus 1
| Supramolecule | Name: Rosellinia necatrix quadrivirus 1 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1000373 / Sci species name: Rosellinia necatrix quadrivirus 1 / Sci species strain: W1118 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Rosellinia necatrix (fungus) Rosellinia necatrix (fungus) |
| Molecular weight | Experimental: 15.9 MDa |
| Virus shell | Shell ID: 1 / Name: P2 and P4 / Diameter: 470.0 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 / Details: 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 5 mM EDTA |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Digitization - Frames/image: 4-21 / Number real images: 1125 / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.7 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-5nd1: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















