-Search query
-Search result
Showing all 34 items for (author: ginger & l)

EMDB-47333: 
CryoEM structure of the human antibodies PIV3HN-05 and PIV3HN-13 in complex with the parainfluenza virus hemagglutinin-neuraminidase protein
Method: single particle / : Mousa JJ, Miller RJ

PDB-9dzq: 
CryoEM structure of the human antibodies PIV3HN-05 and PIV3HN-13 in complex with the parainfluenza virus hemagglutinin-neuraminidase protein
Method: single particle / : Mousa JJ, Miller RJ

EMDB-27031: 
Accurate computational design of genetically encoded 3D protein crystals
Method: single particle / : Li Z, Borst AJ, Baker D

EMDB-40926: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Method: single particle / : Weidle C, Borst A

PDB-8cwy: 
Accurate computational design of genetically encoded 3D protein crystals
Method: single particle / : Li Z, Borst AJ, Baker D

PDB-8szz: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4
Method: single particle / : Weidle C, Borst A

EMDB-26363: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM459
Method: single particle / : Kirk NS, Lawrence MC

EMDB-26364: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462
Method: single particle / : Kirk NS, Lawrence MC

PDB-7u6d: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM459
Method: single particle / : Kirk NS, Lawrence MC

PDB-7u6e: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462
Method: single particle / : Kirk NS, Lawrence MC

EMDB-26704: 
Cryo-EM Structure of the Neutralizing Antibody MPV467 in Complex with Prefusion Human Metapneumovirus F Glycoprotein
Method: single particle / : Rush SA, McLellan JS

PDB-7ur4: 
Cryo-EM Structure of the Neutralizing Antibody MPV467 in Complex with Prefusion Human Metapneumovirus F Glycoprotein
Method: single particle / : Rush SA, McLellan JS

EMDB-26586: 
Negative stain EM map of Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05
Method: single particle / : Han J, Dzimianski JV, DuBois RM, Ward A

EMDB-23211: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

EMDB-23215: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

PDB-7l7f: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

PDB-7l7k: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

EMDB-11628: 
Distance-dependent synaptic vesicle protein organisation.
Method: electron tomography / : Ginger L, Malsam J, Sonnen AF-P, Morado D, Scheutzow A, Sollner TH, Briggs JAG

EMDB-10134: 
CryoEM structure of murine perforin-2 ectodomain in a pre-pore form
Method: single particle / : Ni T, Yu X

EMDB-10135: 
CryoEM structure of murine perforin-2 ectodomain in a pore form
Method: single particle / : Ni T, Yu X

PDB-6sb3: 
CryoEM structure of murine perforin-2 ectodomain in a pre-pore form
Method: single particle / : Ni T, Yu X, Gilbert RJC

PDB-6sb5: 
CryoEM structure of murine perforin-2 ectodomain in a pore form
Method: single particle / : Ni T, Yu X, Gilbert RJC

EMDB-0553: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2
Method: single particle / : Meyer NL, Xie Q

PDB-6nz0: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2
Method: single particle / : Meyer NL, Xie Q, Davulcu O, Yoshioka C, Chapman MS

EMDB-0621: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: subtomogram averaging / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L
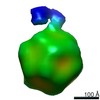
EMDB-0622: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: subtomogram averaging / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L

EMDB-0623: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: electron tomography / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L

EMDB-0624: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: electron tomography / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L

EMDB-7793: 
Thermostablilized dephosphorylated chicken CFTR
Method: single particle / : Fay JF, Riordan JR

EMDB-7794: 
Thermostabilized phosphorylated chicken CFTR
Method: single particle / : Fay JF, Riordan JR

PDB-6d3r: 
Thermostablilized dephosphorylated chicken CFTR
Method: single particle / : Fay JF, Riordan JR

PDB-6d3s: 
Thermostabilized phosphorylated chicken CFTR
Method: single particle / : Fay JF, Riordan JR, Chen ZJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
