[English] 日本語
 Yorodumi
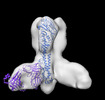
Yorodumi- EMDB-26586: Negative stain EM map of Y2 COBRA hemagglutinin in complex with m... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05 | |||||||||
 Map data Map data | Negative stain EM map of Y2 COBRA hemagglutinin binding monoclonal antibody P1-05 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 11.0 Å | |||||||||
 Authors Authors | Han J / Dzimianski JV / DuBois RM / Ward A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Immunol / Year: 2022 Journal: J Immunol / Year: 2022Title: The Pre-Existing Human Antibody Repertoire to Computationally Optimized Influenza H1 Hemagglutinin Vaccines. Authors: Kaito Nagashima / John V Dzimianski / Julianna Han / Nada Abbadi / Aaron D Gingerich / Fredejah Royer / Sara O'Rourke / Giuseppe A Sautto / Ted M Ross / Andrew B Ward / Rebecca M DuBois / Jarrod J Mousa /  Abstract: Computationally optimized broadly reactive Ag (COBRA) hemagglutinin (HA) immunogens have previously been generated for several influenza subtypes to improve vaccine-elicited Ab breadth. As nearly all ...Computationally optimized broadly reactive Ag (COBRA) hemagglutinin (HA) immunogens have previously been generated for several influenza subtypes to improve vaccine-elicited Ab breadth. As nearly all individuals have pre-existing immunity to influenza viruses, influenza-specific memory B cells will likely be recalled upon COBRA HA vaccination. We determined the epitope specificity and repertoire characteristics of pre-existing human B cells to H1 COBRA HA Ags. Cross-reactivity between wild-type HA and H1 COBRA HA proteins P1, X6, and Y2 were observed for isolated mAbs. The mAbs bound five distinct epitopes on the pandemic A/California/04/2009 HA head and stem domains, and most mAbs had hemagglutination inhibition and neutralizing activity against 2009 pandemic H1 strains. Two head-directed mAbs, CA09-26 and CA09-45, had hemagglutination inhibition and neutralizing activity against a prepandemic H1 strain. One mAb, P1-05, targeted the stem region of H1 HA, but did not compete with a known stem-targeting H1 mAb. We determined that mAb P1-05 recognizes a recently discovered HA epitope, the anchor epitope, and we identified similar mAbs using B cell repertoire sequencing. In addition, the trimerization domain distance from HA was critical to recognition of this epitope by mAb P1-05, suggesting the importance of protein design for vaccine formulations. Overall, these data indicate that seasonally vaccinated individuals possess a population of functional H1 COBRA HA-reactive B cells that target head, central stalk, and anchor epitopes, and they demonstrate the importance of structure-based assessment of subunit protein vaccine candidates to ensure accessibility of optimal protein epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26586.map.gz emd_26586.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26586-v30.xml emd-26586-v30.xml emd-26586.xml emd-26586.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26586.png emd_26586.png | 74.7 KB | ||
| Others |  emd_26586_half_map_1.map.gz emd_26586_half_map_1.map.gz emd_26586_half_map_2.map.gz emd_26586_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26586 http://ftp.pdbj.org/pub/emdb/structures/EMD-26586 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26586 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26586 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26586.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26586.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM map of Y2 COBRA hemagglutinin binding monoclonal antibody P1-05 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.98 Å | ||||||||||||||||||||||||||||||||||||
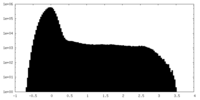
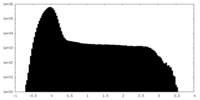
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Negative stain EM half map A of Y2...
| File | emd_26586_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM half map A of Y2 COBRA hemagglutinin binding monoclonal antibody P1-05 | ||||||||||||
| Projections & Slices |
| ||||||||||||
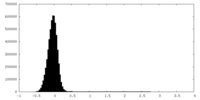
| Density Histograms |
-Half map: Negative stain EM half map B of Y2...
| File | emd_26586_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM half map B of Y2 COBRA hemagglutinin binding monoclonal antibody P1-05 | ||||||||||||
| Projections & Slices |
| ||||||||||||
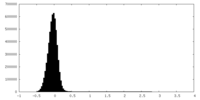
| Density Histograms |
- Sample components
Sample components
-Entire : Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05
| Entire | Name: Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05 |
|---|---|
| Components |
|
-Supramolecule #1: Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05
| Supramolecule | Name: Y2 COBRA hemagglutinin in complex with monoclonal antibody P1-05 type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #2: Y2 COBRA hemagglutinin
| Supramolecule | Name: Y2 COBRA hemagglutinin / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Details: HA antigen based on A/California/04/2009 H1N1 HA |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: P1-05 monoclonal antibody
| Supramolecule | Name: P1-05 monoclonal antibody / type: complex / Chimera: Yes / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl formate / Details: 2% w/v |
| Grid | Model: Homemade / Pretreatment - Type: PLASMA CLEANING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.0 µm / Nominal defocus min: -1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 11.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 12717 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)