+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11628 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Distance-dependent synaptic vesicle protein organisation. | |||||||||||||||
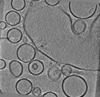
 Map data Map data | Tomogram from delta-84 Munc-18 condition. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | electron tomography / cryo EM | |||||||||||||||
 Authors Authors | Ginger L / Malsam J / Sonnen AF-P / Morado D / Scheutzow A / Sollner TH / Briggs JAG | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: FEBS Lett / Year: 2020 Journal: FEBS Lett / Year: 2020Title: Arrangements of proteins at reconstituted synaptic vesicle fusion sites depend on membrane separation. Authors: Lucy Ginger / Joerg Malsam / Andreas F-P Sonnen / Dustin Morado / Andrea Scheutzow / Thomas H Söllner / John A G Briggs /   Abstract: Synaptic vesicle proteins, including N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), Synaptotagmin-1 and Complexin, are responsible for controlling the synchronised fusion of ...Synaptic vesicle proteins, including N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), Synaptotagmin-1 and Complexin, are responsible for controlling the synchronised fusion of synaptic vesicles with the presynaptic plasma membrane in response to elevated cytosolic calcium levels. A range of structures of SNAREs and their regulatory proteins have been elucidated, but the exact organisation of these proteins at synaptic junction membranes remains elusive. Here, we have used cryoelectron tomography to investigate the arrangement of synaptic proteins in an in vitro reconstituted fusion system. We found that the separation between vesicle and target membranes strongly correlates with the organisation of protein complexes at junctions. At larger membrane separations, protein complexes assume a 'clustered' distribution at the docking site, inducing a protrusion in the target membrane. As the membrane separation decreases, protein complexes become displaced radially outwards and assume a 'ring-like' arrangement. Our findings indicate that docked vesicles can possess a wide range of protein complex numbers and be heterogeneous in their protein arrangements. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11628.map.gz emd_11628.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11628-v30.xml emd-11628-v30.xml emd-11628.xml emd-11628.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11628.png emd_11628.png | 142.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11628 http://ftp.pdbj.org/pub/emdb/structures/EMD-11628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11628 | HTTPS FTP |
-Related structure data
| EM raw data |  EMPIAR-10498 (Title: Arrangements of proteins at reconstituted synaptic vesicle fusion sites depend on membrane separation. EMPIAR-10498 (Title: Arrangements of proteins at reconstituted synaptic vesicle fusion sites depend on membrane separation.Data size: 70.0 Data #1: Tomograms from the delta84 + Munc-18 condition and associated coordinate file. [reconstructed volumes] Data #2: Tomograms from the delta84 condition (collection 1) and associated coordinate file. [reconstructed volumes] Data #3: Tomograms from the delta84 condition (collection 2) and associated coordinate file. [reconstructed volumes] Data #4: Tomograms from the WT condition and associated coordinate file. [reconstructed volumes]) |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11628.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11628.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomogram from delta-84 Munc-18 condition. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.356 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Synaptic membrane fusion reconstituted in vitro
| Entire | Name: Synaptic membrane fusion reconstituted in vitro |
|---|---|
| Components |
|
-Supramolecule #1: Synaptic membrane fusion reconstituted in vitro
| Supramolecule | Name: Synaptic membrane fusion reconstituted in vitro / type: complex / ID: 1 / Parent: 0 Details: Synaptic membrane fusion reconstituted in vitro by mixing: giant unilamellar vesicles reconstituted with t-SNARE complex proteins; small unilamellar vesicles reconstituted with v-SNARE and ...Details: Synaptic membrane fusion reconstituted in vitro by mixing: giant unilamellar vesicles reconstituted with t-SNARE complex proteins; small unilamellar vesicles reconstituted with v-SNARE and Synaptotagmin-1; Complexin-II and Munc-18. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: BBI Solutions / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Number images used: 121 |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)