-Search query
-Search result
Showing 1 - 50 of 118 items for (author: choi & sh)

EMDB entry, No image
EMDB-41849: 
Structure of 310-18A5 Fab in complex with A/Solomon Islands/3/2006(H1N1) influenza virus hemagglutinin

EMDB-37593: 
Vibrio vulnificus MARTX effector duet (RDTND-RID) complexed with human Rac1 Q61L and calmodulin

EMDB-39534: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH
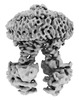
EMDB-39535: 
Cryo-EM structure of the human ABCB6 in complex with Cd(II):Phytochelatin 2

EMDB-40699: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate

EMDB-40707: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode

EMDB-40708: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate

PDB-8sq9: 
SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate

PDB-8sqj: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode

PDB-8sqk: 
SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate

EMDB-29013: 
96nm doublet microtubule repeat from wild type mouse sperm

EMDB-28606: 
96nm doublet microtubule repeat from LRRC23-KO mouse sperm

EMDB-36937: 
post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound)

EMDB-36938: 
Outward-facing structure of human ABCB6 W546A mutant (ADP/VO4-bound)

EMDB-33734: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody

PDB-7yc5: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody

EMDB-34648: 
CryoEM structure of Helicobacter pylori UreFD/urease complex

EMDB-34659: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex

PDB-8hc1: 
CryoEM structure of Helicobacter pylori UreFD/urease complex

PDB-8hcn: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex

EMDB-34350: 
Legionella pneumophila Deubiquitinase, LotA(7-544) apo state

EMDB-27777: 
Streptomyces venezuelae RNAP transcription open promoter complex with WhiA and WhiB transcription factors

EMDB-27778: 
Streptomyces venezuelae RNAP unconstrained open promoter complex with WhiA and WhiB transcription factors

PDB-8dy7: 
Streptomyces venezuelae RNAP transcription open promoter complex with WhiA and WhiB transcription factors

PDB-8dy9: 
Streptomyces venezuelae RNAP unconstrained open promoter complex with WhiA and WhiB transcription factors

EMDB-27254: 
SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196

EMDB-27255: 
SARS-CoV-2 Spike RBD in complex with DMAb 2196

PDB-8d8q: 
SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196

PDB-8d8r: 
SARS-CoV-2 Spike RBD in complex with DMAb 2196

EMDB-31722: 
Cryo-EM structure of the mouse ABCB9 (ADP.BeF3-bound)

EMDB-31723: 
Cryo-EM structure of the mouse ABCB9 (PG-bound)
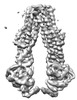
EMDB-31955: 
Cryo-EM structure of the mouse TAPL (9mer-peptide bound)

EMDB-30790: 
Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound)

EMDB-30791: 
Cryo-EM structure of the human ABCB6 (Hemin and GSH-bound)

EMDB-25879: 
Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens

PDB-7tfs: 
Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens

EMDB-32629: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution

EMDB-32630: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution

EMDB-25188: 
Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E)

EMDB-25189: 
Full-length insulin receptor bound with site 2 binding deficient mutant insulin (A-L13R) -- asymmetric conformation

EMDB-25190: 
Full-length insulin receptor bound with site 2 binding deficient mutant insulin (A-L13R) -- symmetric conformation

EMDB-25191: 
Full-length insulin receptor bound with site 2 binding deficient mutant insulin (B-L17R) -- asymmetric conformation

EMDB-25192: 
Full-length insulin receptor bound with site 2 binding deficient mutant insulin (B-L17R) -- symmetric conformation

EMDB-25193: 
Full-length insulin receptor bound with both site 1 binding deficient mutant insulin (A-V3E) and site 2 binding deficient mutant insulin (A-L13R)

EMDB-25428: 
Full-length insulin receptor bound with unsaturated insulin WT (2 insulin bound) symmetric conformation

EMDB-25429: 
Full-length insulin receptor bound with unsaturated insulin WT (1 insulin bound) asymmetric conformation

EMDB-25430: 
Full-length insulin receptor bound with unsaturated insulin WT (2 insulins bound) asymmetric conformation (Conformation 1)

EMDB-25431: 
Full-length insulin receptor bound with unsaturated insulin WT (2 insulins bound) asymmetric conformation (Conformation 2)

PDB-7sl1: 
Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E)

PDB-7sl2: 
Full-length insulin receptor bound with site 2 binding deficient mutant insulin (A-L13R) -- asymmetric conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
