[English] 日本語
 Yorodumi
Yorodumi- EMDB-25188: Full-length insulin receptor bound with site 1 binding deficient ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
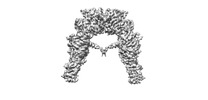
| Title | Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E) | |||||||||
 Map data Map data | Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | insulin receptor / site 1 binding deficient mutant insulin / SIGNALING PROTEIN / SIGNALING PROTEIN-HORMONE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by Insulin receptor / IRS activation / Insulin receptor signalling cascade / Signal attenuation / Insulin receptor recycling / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / regulation of female gonad development / positive regulation of meiotic cell cycle / insulin-like growth factor II binding / positive regulation of developmental growth ...Signaling by Insulin receptor / IRS activation / Insulin receptor signalling cascade / Signal attenuation / Insulin receptor recycling / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / regulation of female gonad development / positive regulation of meiotic cell cycle / insulin-like growth factor II binding / positive regulation of developmental growth / male sex determination / nuclear lumen / insulin receptor complex / insulin-like growth factor I binding / insulin receptor activity / exocrine pancreas development / insulin binding / adrenal gland development / negative regulation of glycogen catabolic process / PTB domain binding / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / Insulin processing / regulation of protein secretion / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / Regulation of gene expression in beta cells / negative regulation of acute inflammatory response / alpha-beta T cell activation / insulin receptor substrate binding / positive regulation of receptor internalization / regulation of embryonic development / Synthesis, secretion, and deacylation of Ghrelin / positive regulation of dendritic spine maintenance / epidermis development / negative regulation of protein secretion / negative regulation of gluconeogenesis / positive regulation of glycogen biosynthetic process / fatty acid homeostasis / Signal attenuation / protein kinase activator activity / positive regulation of insulin receptor signaling pathway / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / negative regulation of respiratory burst involved in inflammatory response / negative regulation of lipid catabolic process / positive regulation of lipid biosynthetic process / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / heart morphogenesis / regulation of protein localization to plasma membrane / phosphatidylinositol 3-kinase binding / transport vesicle / nitric oxide-cGMP-mediated signaling / positive regulation of nitric-oxide synthase activity / COPI-mediated anterograde transport / Insulin receptor recycling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of brown fat cell differentiation / insulin-like growth factor receptor binding / NPAS4 regulates expression of target genes / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / positive regulation of glycolytic process / animal organ morphogenesis / positive regulation of cytokine production / endosome lumen / acute-phase response / positive regulation of long-term synaptic potentiation / positive regulation of D-glucose import across plasma membrane / positive regulation of protein secretion / insulin receptor binding / positive regulation of cell differentiation / Regulation of insulin secretion / wound healing / positive regulation of neuron projection development / hormone activity / receptor protein-tyrosine kinase / negative regulation of protein catabolic process / regulation of synaptic plasticity / caveola / receptor internalization / positive regulation of protein localization to nucleus / Golgi lumen / cellular response to growth factor stimulus / vasodilation / male gonad development / cognition / recycling endosome membrane / glucose metabolic process / positive regulation of nitric oxide biosynthetic process / insulin receptor signaling pathway / late endosome / nuclear envelope / cell-cell signaling / glucose homeostasis / regulation of protein localization Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Bai XC / Choi E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Synergistic activation of the insulin receptor via two distinct sites. Authors: Jie Li / Junhee Park / John P Mayer / Kristofor J Webb / Emiko Uchikawa / Jiayi Wu / Shun Liu / Xuewu Zhang / Michael H B Stowell / Eunhee Choi / Xiao-Chen Bai /  Abstract: Insulin receptor (IR) signaling controls multiple facets of animal physiology. Maximally four insulins bind to IR at two distinct sites, termed site-1 and site-2. However, the precise functional ...Insulin receptor (IR) signaling controls multiple facets of animal physiology. Maximally four insulins bind to IR at two distinct sites, termed site-1 and site-2. However, the precise functional roles of each binding event during IR activation remain unresolved. Here, we showed that IR incompletely saturated with insulin predominantly forms an asymmetric conformation and exhibits partial activation. IR with one insulin bound adopts a Γ-shaped conformation. IR with two insulins bound assumes a Ƭ-shaped conformation. One insulin binds at site-1 and another simultaneously contacts both site-1 and site-2 in the Ƭ-shaped IR dimer. We further show that concurrent binding of four insulins to sites-1 and -2 prevents the formation of asymmetric IR and promotes the T-shaped symmetric, fully active state. Collectively, our results demonstrate how the synergistic binding of multiple insulins promotes optimal IR activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25188.map.gz emd_25188.map.gz | 165.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25188-v30.xml emd-25188-v30.xml emd-25188.xml emd-25188.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25188.png emd_25188.png | 33 KB | ||
| Filedesc metadata |  emd-25188.cif.gz emd-25188.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25188 http://ftp.pdbj.org/pub/emdb/structures/EMD-25188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25188 | HTTPS FTP |
-Related structure data
| Related structure data |  7sl1MC  7sl2C  7sl3C  7sl4C  7sl6C  7sl7C  7sthC  7stiC  7stjC  7stkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25188.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25188.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Full-length insulin receptor bound with site 1 binding deficient ...
| Entire | Name: Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E) |
|---|---|
| Components |
|
-Supramolecule #1: Full-length insulin receptor bound with site 1 binding deficient ...
| Supramolecule | Name: Full-length insulin receptor bound with site 1 binding deficient mutant insulin (A-V3E) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Macromolecule #1: Insulin receptor
| Macromolecule | Name: Insulin receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 155.790516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGFGRGCETT AVPLLVAVAA LLVGTAGHLY PGEVCPGMDI RNNLTRLHEL ENCSVIEGHL QILLMFKTRP EDFRDLSFPK LIMITDYLL LFRVYGLESL KDLFPNLTVI RGSRLFFNYA LVIFEMVHLK ELGLYNLMNI TRGSVRIEKN NELCYLATID W SRILDSVE ...String: MGFGRGCETT AVPLLVAVAA LLVGTAGHLY PGEVCPGMDI RNNLTRLHEL ENCSVIEGHL QILLMFKTRP EDFRDLSFPK LIMITDYLL LFRVYGLESL KDLFPNLTVI RGSRLFFNYA LVIFEMVHLK ELGLYNLMNI TRGSVRIEKN NELCYLATID W SRILDSVE DNYIVLNKDD NEECGDVCPG TAKGKTNCPA TVINGQFVER CWTHSHCQKV CPTICKSHGC TAEGLCCHKE CL GNCSEPD DPTKCVACRN FYLDGQCVET CPPPYYHFQD WRCVNFSFCQ DLHFKCRNSR KPGCHQYVIH NNKCIPECPS GYT MNSSNL MCTPCLGPCP KVCQILEGEK TIDSVTSAQE LRGCTVINGS LIINIRGGNN LAAELEANLG LIEEISGFLK IRRS YALVS LSFFRKLHLI RGETLEIGNY SFYALDNQNL RQLWDWSKHN LTITQGKLFF HYNPKLCLSE IHKMEEVSGT KGRQE RNDI ALKTNGDQAS CENELLKFSF IRTSFDKILL RWEPYWPPDF RDLLGFMLFY KEAPYQNVTE FDGQDACGSN SWTVVD IDP PQRSNDPKSQ TPSHPGWLMR GLKPWTQYAI FVKTLVTFSD ERRTYGAKSD IIYVQTDATN PSVPLDPISV SNSSSQI IL KWKPPSDPNG NITHYLVYWE RQAEDSELFE LDYCLKGLKL PSRTWSPPFE SDDSQKHNQS EYDDSASECC SCPKTDSQ I LKELEESSFR KTFEDYLHNV VFVPRPSRKR RSLEEVGNVT ATTLTLPDFP NVSSTIVPTS QEEHRPFEKV VNKESLVIS GLRHFTGYRI ELQACNQDSP DERCSVAAYV SARTMPEAKA DDIVGPVTHE IFENNVVHLM WQEPKEPNGL IVLYEVSYRR YGDEELHLC VSRKHFALER GCRLRGLSPG NYSVRVRATS LAGNGSWTEP TYFYVTDYLD VPSNIAKIII GPLIFVFLFS V VIGSIYLF LRKRQPDGPM GPLYASSNPE YLSASDVFPS SVYVPDEWEV PREKITLLRE LGQGSFGMVY EGNAKDIIKG EA ETRVAVK TVNESASLRE RIEFLNEASV MKGFTCHHVV RLLGVVSKGQ PTLVVMELMA HGDLKSHLRS LRPDAENNPG RPP PTLQEM IQMTAEIADG MAYLNAKKFV HRDLAARNCM VAHDFTVKIG DFGMTRDIYE TDYYRKGGKG LLPVRWMSPE SLKD GVFTA SSDMWSFGVV LWEITSLAEQ PYQGLSNEQV LKFVMDGGYL DPPDNCPERL TDLMRMCWQF NPKMRPTFLE IVNLL KDDL HPSFPEVSFF YSEENKAPES EELEMEFEDM ENVPLDRSSH CQREEAGGRE GGSSLSIKRT YDEHIPYTHM NGGKKN GRV LTLPRSNPS UniProtKB: Insulin receptor |
-Macromolecule #2: Insulin B chain
| Macromolecule | Name: Insulin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.433953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FVNQHLCGSH LVEALYLVCG ERGFFYTPKT UniProtKB: Insulin |
-Macromolecule #3: Insulin A chain
| Macromolecule | Name: Insulin A chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.413681 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GIEEQCCTSI CSLYQLENYC N UniProtKB: Insulin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















