-Search query
-Search result
Showing all 41 items for (author: calero & g)

EMDB-43114: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43115: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43116: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43117: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43120: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43121: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43122: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43123: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43124: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43125: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

EMDB-43126: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vb6: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vb7: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vb8: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vb9: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbc: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbd: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbe: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbf: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbg: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbh: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G

PDB-8vbi: 
Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM
Method: single particle / : Vergara S, Zhou X, Santiago U, Conway JF, Sluis-Cremer N, Calero G
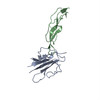
PDB-5ty4: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ, Hattne J, Shi D, Reyes FE, Callero G, Gonen T

PDB-5k7n: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7o: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7p: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7q: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7r: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7s: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7t: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8216: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8217: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8218: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8219: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8220: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8221: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8222: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8472: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ

PDB-3j1n: 
Cryo-EM map of a yeast minimal preinitiation complex interacting with the Mediator Head module
Method: single particle / : Asturias FJ, Imasaki T

PDB-3j1o: 
Cryo-EM map of a yeast minimal preinitiation complex interacting with the Mediator Head module
Method: single particle / : Asturias FJ, Imasaki T

EMDB-5407: 
Cryo-EM map of a yeast minimal preinitiation complex interacting with the Mediator Head module
Method: single particle / : Cai G, Chaban Y, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias FJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
