-Search query
-Search result
Showing all 34 items for (author: bornholdt & za)

EMDB-23488: 
Hendra virus receptor binding protein in complex with HENV-103 and HENV-117 Fabs

EMDB-22884: 
Structure of the NiV F glycoprotein in complex with the 12B2 neutralizing antibody

EMDB-22885: 
Structure of the HeV F glycoprotein in complex with the 1F5 neutralizing antibody

PDB-7ki4: 
Structure of the NiV F glycoprotein in complex with the 12B2 neutralizing antibody

PDB-7ki6: 
Structure of the HeV F glycoprotein in complex with the 1F5 neutralizing antibody

EMDB-11660: 
Structure of VP40 matrix layer in EBOV NP-VP24-VP35-VP40 VLPs

EMDB-11661: 
Structure of VP40 matrix layer in EBOV VP40 VLPs

EMDB-11662: 
Structure of VP40 matrix layer in EBOV VP40-GP VLPs

EMDB-11663: 
Structure of VP40 matrix layer in intact MARV virions

EMDB-11664: 
Structure of VP40 matrix layer in MARV VP40 VLPs

EMDB-11665: 
Structure of GP in EBOV VP40-GP VLPs

EMDB-8935: 
Ebola virus Makona variant GP (mucin-deleted) in complex with pan-ebolavirus human antibody ADI-15878 Fab

EMDB-8936: 
Bundibugyo virus GP (mucin-deleted) in complex with pan-ebolavirus human antibody ADI-15878 Fab

PDB-6dzl: 
Ebola virus Makona variant GP (mucin-deleted) in complex with pan-ebolavirus human antibody ADI-15878 Fab

PDB-6dzm: 
Bundibugyo virus GP (mucin-deleted) in complex with pan-ebolavirus human antibody ADI-15878 Fab
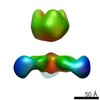
EMDB-8698: 
EBOV GPdMuc:ADI-16061

EMDB-8699: 
EBOV GPdMuc in complex with ADI-15742 Fab

EMDB-8700: 
EBOV GPdMuc:ADI-15878

EMDB-8701: 
EBOV GPdMuc:ADI-15946

EMDB-6586: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies

EMDB-6587: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
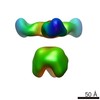
EMDB-6588: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
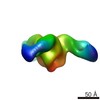
EMDB-6589: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies

EMDB-6232: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor

EMDB-6233: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor

EMDB-6234: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor

EMDB-6235: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor
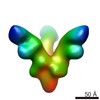
EMDB-6236: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor

EMDB-6237: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor

EMDB-6238: 
Negative stain single particle electron microscopy of Marburg Virus Glycoproteins bound to antibodies from a human survivor
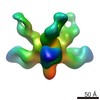
EMDB-6150: 
Structures of Protective Antibodies Reveal Sites of Vulnerability on Ebola Virus
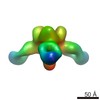
EMDB-6151: 
Structures of Protective Antibodies Reveal Sites of Vulnerability on Ebola Virus
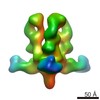
EMDB-6152: 
Structures of Protective Antibodies Reveal Sites of Vulnerability on Ebola Virus
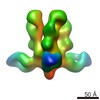
EMDB-6153: 
Structures of Protective Antibodies Reveal Sites of Vulnerability on Ebola Virus
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
