-Search query
-Search result
Showing 1 - 50 of 79 items for (author: bloom & jd)

EMDB-46708: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. Complex of fAPN with FCoV-23 RBD

EMDB-46709: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. FCoV-23 S short

EMDB-46710: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. FCoV-23 S Do in proximal conformation (local refinement)

EMDB-46714: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. FCoV-23 S long with Do in swung-out conformation

EMDB-46716: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. FCoV-23 S long domain 0 in swung-out conformation (local refinement)

EMDB-46739: 
Molecular basis of pathogenicity of the recently emerged FCoV-23 coronavirus. FCoV-23 S long with Do in mixed conformations (global refinement).

EMDB-70353: 
Clone 2.1 Fab in complex with chicken IgY CH2 domain (local refinement)

EMDB-44627: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1533 (local refinement of NTD and C1533)

EMDB-44628: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596

EMDB-44629: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952

EMDB-43250: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, global refinement

EMDB-43261: 
SARS-CoV-2 spike omicron (BA.1) ectodomain dimer-of-trimers in complex with SC27 Fab, global refinement

EMDB-43260: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement

EMDB-43315: 
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs

EMDB-16375: 
SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody

EMDB-42970: 
Model and map from local refinement of a CAB-A17 - Omicron Ba.1 spike complex

EMDB-16397: 
SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody

EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

EMDB-26583: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)

EMDB-29052: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 3 - disordered)

EMDB-29053: 
SARS-CoV-2 Spike Hexapro - C59.68 Fab (Class 1 - No Fab bound)

EMDB-29054: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 2 - Fab bound)

EMDB-26727: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation

EMDB-26729: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)

EMDB-26730: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation

EMDB-26731: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175

EMDB-25785: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (three receptor-binding domains open)

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA

EMDB-25783: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (local refinement of the RBD and S2K146)

EMDB-25784: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (two receptor-binding domains open)

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
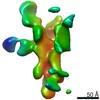
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
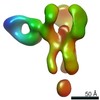
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
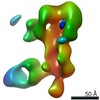
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
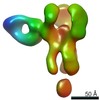
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
