[English] 日本語
 Yorodumi
Yorodumi- EMDB-26727: Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
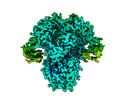
| Title | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human coronavirus / Coronavirus / CCoV-HuPn-2018 / spike glycoprotein / Alpha-coronaviruses / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  unidentified human coronavirus unidentified human coronavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Tortorici MA / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein. Authors: M Alejandra Tortorici / Alexandra C Walls / Anshu Joshi / Young-Jun Park / Rachel T Eguia / Marcos C Miranda / Elizabeth Kepl / Annie Dosey / Terry Stevens-Ayers / Michael J Boeckh / Amalio ...Authors: M Alejandra Tortorici / Alexandra C Walls / Anshu Joshi / Young-Jun Park / Rachel T Eguia / Marcos C Miranda / Elizabeth Kepl / Annie Dosey / Terry Stevens-Ayers / Michael J Boeckh / Amalio Telenti / Antonio Lanzavecchia / Neil P King / Davide Corti / Jesse D Bloom / David Veesler /   Abstract: The isolation of CCoV-HuPn-2018 from a child respiratory swab indicates that more coronaviruses are spilling over to humans than previously appreciated. We determined the structures of the CCoV-HuPn- ...The isolation of CCoV-HuPn-2018 from a child respiratory swab indicates that more coronaviruses are spilling over to humans than previously appreciated. We determined the structures of the CCoV-HuPn-2018 spike glycoprotein trimer in two distinct conformational states and showed that its domain 0 recognizes sialosides. We identified that the CCoV-HuPn-2018 spike binds canine, feline, and porcine aminopeptidase N (APN) orthologs, which serve as entry receptors, and determined the structure of the receptor-binding B domain in complex with canine APN. The introduction of an oligosaccharide at position N739 of human APN renders cells susceptible to CCoV-HuPn-2018 spike-mediated entry, suggesting that single-nucleotide polymorphisms might account for viral detection in some individuals. Human polyclonal plasma antibodies elicited by HCoV-229E infection and a porcine coronavirus monoclonal antibody inhibit CCoV-HuPn-2018 spike-mediated entry, underscoring the cross-neutralizing activity among ɑ-coronaviruses. These data pave the way for vaccine and therapeutic development targeting this zoonotic pathogen representing the eighth human-infecting coronavirus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26727.map.gz emd_26727.map.gz | 483.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26727-v30.xml emd-26727-v30.xml emd-26727.xml emd-26727.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26727.png emd_26727.png | 105.4 KB | ||
| Filedesc metadata |  emd-26727.cif.gz emd-26727.cif.gz | 6.5 KB | ||
| Others |  emd_26727_additional_1.map.gz emd_26727_additional_1.map.gz emd_26727_half_map_1.map.gz emd_26727_half_map_1.map.gz emd_26727_half_map_2.map.gz emd_26727_half_map_2.map.gz | 2 MB 475.1 MB 475.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26727 http://ftp.pdbj.org/pub/emdb/structures/EMD-26727 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26727 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26727 | HTTPS FTP |
-Related structure data
| Related structure data |  7us6MC  7u0lC  7us9C  7usaC  7usbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26727.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26727.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.843 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26727_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26727_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26727_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : unidentified human coronavirus
| Entire | Name:  unidentified human coronavirus unidentified human coronavirus |
|---|---|
| Components |
|
-Supramolecule #1: unidentified human coronavirus
| Supramolecule | Name: unidentified human coronavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 694448 / Sci species name: unidentified human coronavirus / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unidentified human coronavirus unidentified human coronavirus |
| Molecular weight | Theoretical: 142.666391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HWNLIENFLL NYSIRLPPNS DVVLGDYFPT VQPWFNCIRN NNNSLYVTME NLKALYWDYA TENITSDHRQ PVYDVTYYRV NNKNGTTIV SNCTDQCASY VDNVFTTQPG GLIPSDFSFN NWFLLTNSST VVSGKLVTRQ PLVVNCLWPV PSFKEAASTF C FEGAGFDQ ...String: HWNLIENFLL NYSIRLPPNS DVVLGDYFPT VQPWFNCIRN NNNSLYVTME NLKALYWDYA TENITSDHRQ PVYDVTYYRV NNKNGTTIV SNCTDQCASY VDNVFTTQPG GLIPSDFSFN NWFLLTNSST VVSGKLVTRQ PLVVNCLWPV PSFKEAASTF C FEGAGFDQ CNGAVLNNTV DVIRFNLNFT ADVQSGMGAT VFSLNTTGGV ILEISCYNDI VSESSFYSYG DIPFGITDGP RY CYVLYNG TTLKYLGTLP PSVKEIAISK WGHFYINGYN FFSTFPIDCI SFNLTTGASG AFWTIAYTSY TEALVQVENT AIK KVTYCN SHINNIKCSQ LTANLQNGFY PVASSEVGLV NKSVVLLPSF FAHTTVNITI DLGMKRSGYG QPIASPLSNI TLPM QDNNT DVYCIRSNQF SIYVHSTCKS SLWDNVFNQD CTDVLEATAV IKTGTCPFSF DKLNNHLTFN KFCLSLSPVG ANCKF DVAA RTRTNEQVVR SLYVIYEEGD NIVGVPSDNS GLHDLSVLHL DSCTDYNIYG RTGVGVIRQT NSTLLSGLYY TSLSGD LLG FKNVSDGVIY SVTPCDVSAQ AAVIDGAIVG AMTSINSELL GLTHWTTTPN FYYYSIYNYT NERTRGTAID SNDVDCE PI ITYSNIGVCK NGALVFINVT HSDGDVQPIS TGNVTIPTNF TISVQVEYIQ VYTTPVSIDC ARYVCNGNPR CNKLLTQY V SACQTIEQAL AMGARLENME VDSMLFVSEN ALKLASVEAF NSTENLDPIY KEWPNIGGSW LGGLKDILPS HNSKRKYRS AIEDLLFDKV VTSGLGTVDE DYKRCTGGYD IADLVCAQYY NGIMVLPGVA NDDKMTMYTA SLAGGITLGA LGGGAVAIPF AVAVQARLN YVALQTDVLN KNQQILANAF NQAIGNITQA FGKVNDAIHQ TSKGLATVAK VLAKVQDVVN TQGQALSHLT V QLQNNFQA ISSSISDIYN RLDPPSADAQ VDRLITGRLT ALNAFVSQTL TRQAEVRASR QLAKDKVNEC VRSQSQRFGF CG NGTHLFS LANAAPNGMI FFHTVLLPTA YQTVTACSGI CASDGDRTFG LVVKDVQLTL FRNLDDKFYL TPRTMYQPRA ATS SDFVQI EGCDVLFVNA TEIDLPSIIP DYIDINQTVQ DILENYRPNW TVPELTLDIF NATYLNLTGE IDDLEFRSEK LHNT TVELA ILIDNINNTL VNLEWLNRIE TYVKSGGYIP EAPRDGQAYV RKDGEWVLLS TFLVPRGSGG SGGSGLNDIF EAQKI EWHE GGSHHHHHHH H UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 43 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Sugar embedding | Material: protein |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 38863 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)