-Search query
-Search result
Showing 1 - 50 of 68 items for (author: robertson & mj)

EMDB-41908: 
Local refinement map on VFT-CRD of active-state CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41909: 
Consensus refinement map of active-state human CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41910: 
Local refinement map on CRD-7TM of active-state CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41925: 
Local refinement map on VFT-CRD of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41926: 
Local refinement map on Gi of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41927: 
Local refinement map on CRD-7TM of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41928: 
Consensus refinement map of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41949: 
Consensus refinement map of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41950: 
Local refinement map on CRD-7TM of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41951: 
Local refinement map on VFT-CRD of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41952: 
Local refinement map on Gq of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41953: 
Consensus refinement map of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41954: 
Local refinement map on VFT-CRD of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41956: 
Local refinement map on CRD-7TM of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-41957: 
Local refinement map on Gi of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-40914: 
Cryo-EM structure of cinacalcet-bound active-state human calcium-sensing receptor CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G
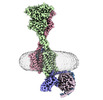
EMDB-40915: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-40916: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G
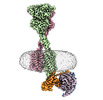
EMDB-40917: 
Cryo-EM structure of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-40052: 
Cannabinoid Receptor 1-G Protein Complex
Method: single particle / : Krishna Kumar K, Robertson MJ, Skiniotis G, Kobilka BK

EMDB-40057: 
Cannabinoid Receptor 1/G protein complex, Global Refinement
Method: single particle / : Krishna Kumar K, Robertson MJ, Skiniotis G, Kobilka BK
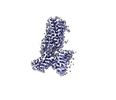
EMDB-40058: 
Cannabinoid Receptor 1/G protein complex, local refinement
Method: single particle / : Krishna Kumar K, Robertson MJ, Skiniotis G, Kobilka BK

EMDB-26313: 
C6-guano bound Mu Opioid Receptor-Gi Protein Complex
Method: single particle / : Wang H, Kobilka B

EMDB-25612: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to mitragynine pseudoindoxyl (MP)
Method: single particle / : Seven AB, Qu Q, Robertson MJ, Wang H, Kobilka BK, Skiniotis G

EMDB-25613: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to lofentanil (LFT)
Method: single particle / : Seven AB, Qu Q, Huang W, Robertson MJ, Kobilka BK, Skiniotis G

EMDB-25401: 
5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-25402: 
5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Cao C, Roth BL, Skiniotis G

EMDB-25403: 
5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-31962: 
The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL
Method: single particle / : Wang XK, Chen G, Liao QW, Du Y, Hu HL, Ye DQ
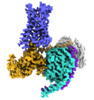
EMDB-24378: 
5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Roth BL, Skiniotis G

EMDB-26589: 
CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6
Method: single particle / : Robertson MJ, Skiniotis G

EMDB-26590: 
CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab
Method: single particle / : Robertson MJ, Skiniotis G

EMDB-26591: 
CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6
Method: single particle / : Robertson MJ, Skiniotis G

EMDB-26592: 
CryoEM Structure of Inactive SSTR2 bound to Nb6
Method: single particle / : Robertson MJ, Skiniotis G

EMDB-31323: 
The structure of formyl peptide receptor 1 in complex with Gi and peptide agonist fMLF
Method: single particle / : Wang XK, Chen G, Liao QW, Du Y, Hu HL, Ye DQ

EMDB-26314: 
C5guano-uOR-Gi-scFv16
Method: single particle / : Wang H, Qu Q, Skiniotis G, Kobilka B

EMDB-25076: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

EMDB-25077: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

EMDB-24733: 
Oxytocin receptor (OTR) bound to oxytocin in complex with a heterotrimeric Gq protein
Method: single particle / : Meyerowitz JG, Panova O, Skiniotis G

EMDB-25586: 
CryoEM structure of somatostatin receptor 2 in complex with somatostatin-14 and Gi3
Method: single particle / : Robertson MJ, Skinotis G

EMDB-25587: 
CryoEM structure of somatostatin receptor 2 in complex with Octreotide and Gi3.
Method: single particle / : Robertson MJ, Skinotis G

EMDB-25194: 
CryoEM structure of SGLT1 at 3.4 A resolution
Method: single particle / : Qu Q, Han L, Panova O, Feng L, Skiniotis G

EMDB-25195: 
CryoEM structure of SMCT1
Method: single particle / : Qu Q, Han L, Panova O, Feng L, Skiniotis G

EMDB-25196: 
CryoEM structure of SGLT1 at 3.15 Angstrom resolution
Method: single particle / : Qu Q, Han L, Panova O, Feng L, Skiniotis G

EMDB-22150: 
CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA

EMDB-22151: 
CryoEM structure of GIRK2-PIP2** - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA

EMDB-22152: 
CryoEM structure of GIRK2-PIP2*** - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA

EMDB-22153: 
CryoEM structure of GIRK2-PIP2**** - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA

EMDB-22154: 
CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA

EMDB-22155: 
CryoEM structure of apo G protein-gated inwardly rectifying potassium channel GIRK2
Method: single particle / : Mathiharan YK, Glaaser IW, Skiniotis G, Slesinger PA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
