[English] 日本語
 Yorodumi
Yorodumi- EMDB-22150: CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectify... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22150 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | |||||||||
 Map data Map data | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GIRK / inwardly rectifying potassium channel / cholesterol / lipids / PIP2 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG-protein activated inward rectifier potassium channel activity / inward rectifier potassium channel complex Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Mathiharan YK / Glaaser IW / Skiniotis G / Slesinger PA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Structural insights into GIRK2 channel modulation by cholesterol and PIP. Authors: Yamuna Kalyani Mathiharan / Ian W Glaaser / Yulin Zhao / Michael J Robertson / Georgios Skiniotis / Paul A Slesinger /  Abstract: G-protein-gated inwardly rectifying potassium (GIRK) channels are important for determining neuronal excitability. In addition to G proteins, GIRK channels are potentiated by membrane cholesterol, ...G-protein-gated inwardly rectifying potassium (GIRK) channels are important for determining neuronal excitability. In addition to G proteins, GIRK channels are potentiated by membrane cholesterol, which is elevated in the brains of people with neurodegenerative diseases such as Alzheimer's dementia and Parkinson's disease. The structural mechanism of cholesterol modulation of GIRK channels is not well understood. In this study, we present cryo- electron microscopy (cryoEM) structures of GIRK2 in the presence and absence of the cholesterol analog cholesteryl hemisuccinate (CHS) and phosphatidylinositol 4,5-bisphosphate (PIP). The structures reveal that CHS binds near PIP in lipid-facing hydrophobic pockets of the transmembrane domain. Our structural analysis suggests that CHS stabilizes PIP interaction with the channel and promotes engagement of the cytoplasmic domain onto the transmembrane region. Mutagenesis of one of the CHS binding pockets eliminates cholesterol-dependent potentiation of GIRK2. Elucidating the structural mechanisms underlying cholesterol modulation of GIRK2 channels could facilitate the development of therapeutics for treating neurological diseases. VIDEO ABSTRACT. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22150.map.gz emd_22150.map.gz | 65.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22150-v30.xml emd-22150-v30.xml emd-22150.xml emd-22150.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
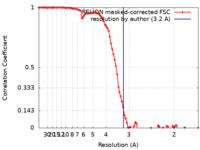
| FSC (resolution estimation) |  emd_22150_fsc.xml emd_22150_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22150.png emd_22150.png | 136.4 KB | ||
| Filedesc metadata |  emd-22150.cif.gz emd-22150.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22150 http://ftp.pdbj.org/pub/emdb/structures/EMD-22150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22150 | HTTPS FTP |
-Related structure data
| Related structure data |  6xeuMC  6xevC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22150.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22150.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : G protein-gated inwardly rectifying potassium channel (GIRK2)
| Entire | Name: G protein-gated inwardly rectifying potassium channel (GIRK2) |
|---|---|
| Components |
|
-Supramolecule #1: G protein-gated inwardly rectifying potassium channel (GIRK2)
| Supramolecule | Name: G protein-gated inwardly rectifying potassium channel (GIRK2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The cryoEM structure obtained in the presence of modulator PIP2. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: G protein-activated inward rectifier potassium channel 2
| Macromolecule | Name: G protein-activated inward rectifier potassium channel 2 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.061957 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MAKRKIQRYV RKDGKCNVHH GNVRETYRYL TDIFTTLVDL KWRFNLLIFV MVYTVTWLFF GMIWWLIAYI RGDMDHIEDP SWTPCVTNL NGFVSAFLFS IETETTIGYG YRVITDKCPE GIILLLIQSV LGSIVNAFMV GCMFVKISQP KKRAETLVFS T HAVISMRD ...String: MAKRKIQRYV RKDGKCNVHH GNVRETYRYL TDIFTTLVDL KWRFNLLIFV MVYTVTWLFF GMIWWLIAYI RGDMDHIEDP SWTPCVTNL NGFVSAFLFS IETETTIGYG YRVITDKCPE GIILLLIQSV LGSIVNAFMV GCMFVKISQP KKRAETLVFS T HAVISMRD GKLCLMFRVG DLRNSHIVEA SIRAKLIKSK QTSEGEFIPL NQTDINVGYY TGDDRLFLVS PLIISHEINQ QS PFWEISK AQLPKEELEI VVILEGMVEA TGMTCQARSS YITSEILWGY RFTPVLTLED GFYEVDYNSF HETYETSTPS LSA KELAEL ANRAESNSLE VLFQ UniProtKB: G protein-activated inward rectifier potassium channel 2 |
-Macromolecule #2: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(o...
| Macromolecule | Name: [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate type: ligand / ID: 2 / Number of copies: 4 / Formula: PIO |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information |  ChemComp-PIO: |
-Macromolecule #3: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 4 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||
| Details | GIRK2PIP2* with the modulator PIP2 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 6480 / Average exposure time: 3.0 sec. / Average electron dose: 83.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)