1ZHO
 
 | | The structure of a ribosomal protein L1 in complex with mRNA | | Descriptor: | 50S ribosomal protein L1, POTASSIUM ION, mRNA | | Authors: | Nevskaya, N, Tishchenko, S, Volchkov, S, Kljashtorny, V, Nikonova, E, Nikonov, O, Nikulin, A, Kohrer, C, Piendl, W, Zimmermann, R, Stockley, P, Garber, M, Nikonov, S. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights into the interaction of ribosomal protein L1 with RNA.
J.Mol.Biol., 355, 2006
|
|
1MZP
 
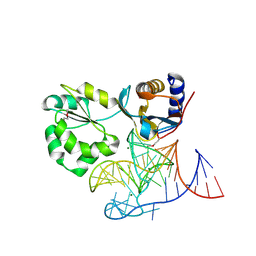 | | Structure of the L1 protuberance in the ribosome | | Descriptor: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | Authors: | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | Deposit date: | 2002-10-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
5A6U
 
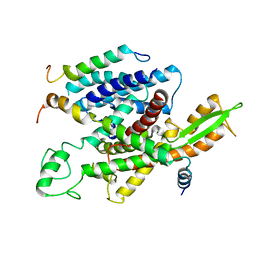 | | Native mammalian ribosome-bound Sec61 protein-conducting channel in the 'non-inserting' state | | Descriptor: | SEC61A, SEC61B, SEC61G | | Authors: | Pfeffer, S, Burbaum, L, Unverdorben, P, Pech, M, Chen, Y, Zimmermann, R, Beckmann, R, Foerster, F. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the Native Sec61 Protein-Conducting Channel.
Nat.Commun., 6, 2015
|
|
6EIC
 
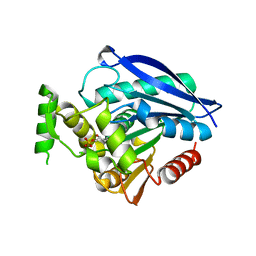 | | Crystal structure of Rv0183, a Monoglyceride Lipase from Mycobacterium Tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycobacterium Tuberculosis Monoglyceride Lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition.
Sci Rep, 8, 2018
|
|
4ZWN
 
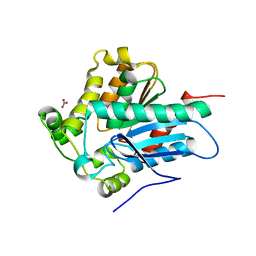 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
8BFO
 
 | | Structure of the apo form of Mpro from SARS-CoV-2 | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
8BGA
 
 | | Structure of Mpro in complex with FGA146 | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
8BFQ
 
 | | Structure of the apo form of Mpro from SARS-CoV-2 | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
8BGD
 
 | | Structure of Mpro from SARS-CoV-2 in complex with FGA147 | | Descriptor: | (phenylmethyl) N-[(2S)-4-methyl-1-[[(2S)-4-nitro-1-[(3R)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
3RLI
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 in complex with PMSF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase, phenylmethanesulfonic acid | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-19 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
3RM3
 
 | | Crystal structure of monoacylglycerol lipase from Bacillus sp. H257 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Bezerra, G.A, Gruber, K, Oberer, M. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes.
Biochim.Biophys.Acta, 1821, 2012
|
|
7ZDU
 
 | |
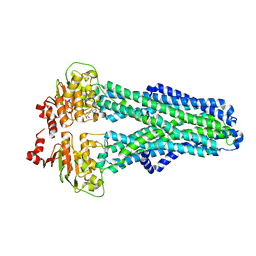
7ZDC
 
 | |
7ZD5
 
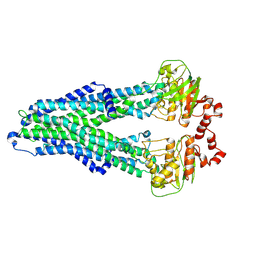 | | IF(apo/as isolated) conformation of CydDC (Dataset-1) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDV
 
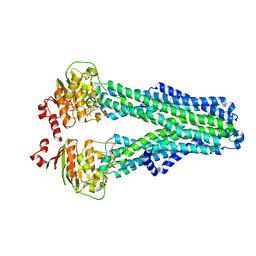 | |
7ZDA
 
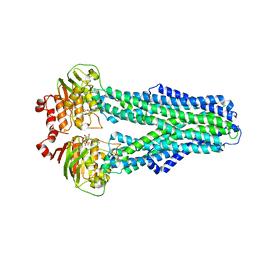 | |
7ZDR
 
 | |
7ZDF
 
 | |
7ZDT
 
 | |
7ZDB
 
 | |
7ZDE
 
 | |
7ZEC
 
 | |
7ZDG
 
 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
