3DXS
 
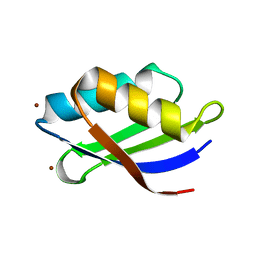 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
2M37
 
 | | Structure of lasso peptide astexin-1 | | Descriptor: | ASTEXIN-1 | | Authors: | Zimmermann, M, Hegemann, J.D, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | The astexin-1 lasso peptides: biosynthesis, stability, and structural studies.
Chem.Biol., 20, 2013
|
|
2MLJ
 
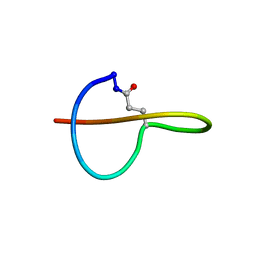 | |
6EQC
 
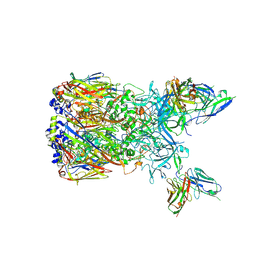 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
8CQU
 
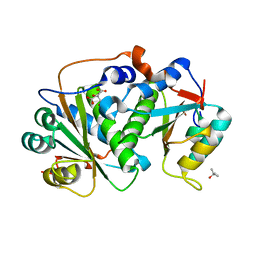 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1680) | | Descriptor: | CITRIC ACID, Putative NADH dehydrogenase/NAD(P)H nitroreductase, TERTIARY-BUTYL ALCOHOL | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQT
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1316) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Putative NADH dehydrogenase/NAD(P)H nitroreductase | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQV
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQS
 
 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_0217) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like protein, PHOSPHATE ION | | Authors: | Blaha, J, Gratzl, S, Mortensen, S.A, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
5OGI
 
 | | Complex of a binding protein and human adenovirus C 5 hexon | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
4NAG
 
 | | Xanthomonins I III are a New Class of Lasso Peptides Featuringa Seven-Membered Macrolactam Ring | | Descriptor: | HEXANE-1,6-DIOL, Xanthomonin I | | Authors: | Hegemann, J.D, Zimmermann, M, Zhu, S, Steuber, H, Harms, K, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-10-22 | | Release date: | 2014-04-30 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Xanthomonins I-III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2MMT
 
 | | Lasso peptide-based integrin inhibitor: Microcin J25 variant with RGDF substitution of Gly12-Ile13-Gly14-Thr15 | | Descriptor: | Microcin J25 RGDF mutant | | Authors: | Hegemann, J.D, Zimmermann, M, Knappe, T.A, Xie, X, Marahiel, M.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-07-02 | | Last modified: | 2014-07-23 | | Method: | SOLUTION NMR | | Cite: | Rational improvement of the affinity and selectivity of integrin binding of grafted lasso peptides.
J.Med.Chem., 57, 2014
|
|
2L41
 
 | | Nab3 RRM - UCUU complex | | Descriptor: | RNA (5'-R(P*UP*CP*UP*U)-3'), RRM domain from Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Stefl, R, Pergoli, R, Hobor, F, Kubicek, K, Zimmermann, M, Pasulka, J, Hofr, C. | | Deposit date: | 2010-09-28 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein
J.Biol.Chem., 286, 2011
|
|
2LX6
 
 | |
2MFV
 
 | |
2KKH
 
 | | Structure of the zinc binding domain of the ATPase HMA4 | | Descriptor: | Putative heavy metal transporter, ZINC ION | | Authors: | Zimmerman, M, Clarke, O, Gulbis, J.M, Keizer, D.W, Jarvis, R.S, Cobbett, C.S, Hinds, M.G, Xiao, Z, Wedd, A.G. | | Deposit date: | 2009-06-20 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains
Biochemistry, 48, 2009
|
|
2L2K
 
 | |
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | Descriptor: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Fraser, J.S, Van Benschoten, A.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
4OBU
 
 | |
3UVQ
 
 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
2L2J
 
 | |
2L3C
 
 | |
2L3J
 
 | |
