2Z61
 
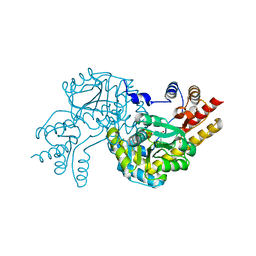 | |
2BBR
 
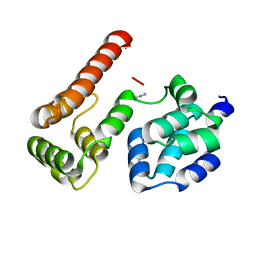 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | AZIDE ION, Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-17 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
2BBZ
 
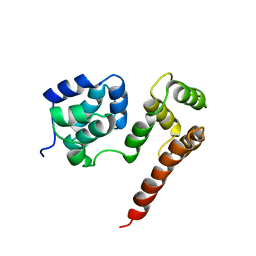 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
1PX8
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1UHV
 
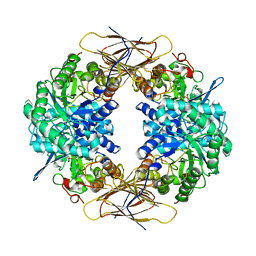 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1NFQ
 
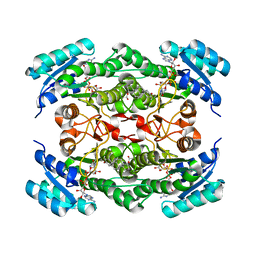 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFF
 
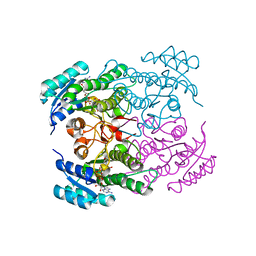 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1C2A
 
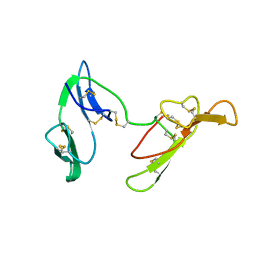 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1EW4
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI CYAY PROTEIN REVEALS A NOVEL FOLD FOR THE FRATAXIN FAMILY | | Descriptor: | CYAY PROTEIN | | Authors: | Suh, S.W, Cho, S, Lee, M.G, Yang, J.K, Lee, J.Y, Song, H.K. | | Deposit date: | 2000-04-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4QB9
 
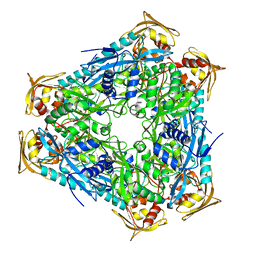 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1UAK
 
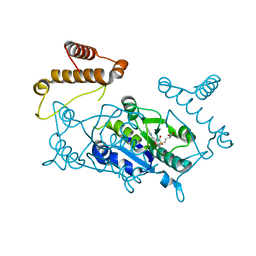 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAL
 
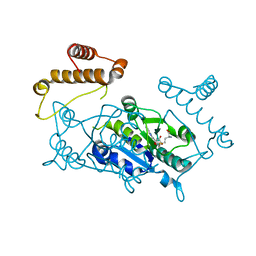 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAJ
 
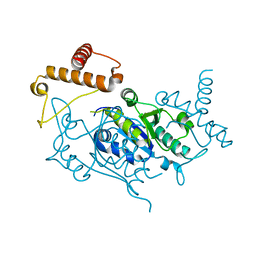 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAM
 
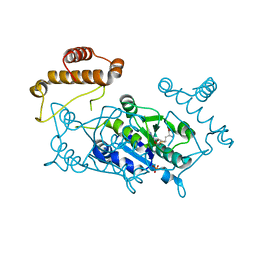 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
5GLF
 
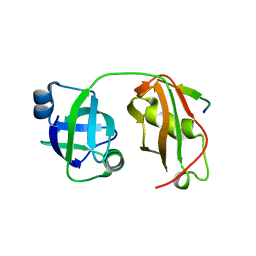 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
5B6C
 
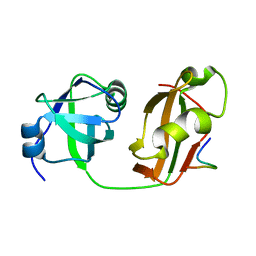 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
8HL6
 
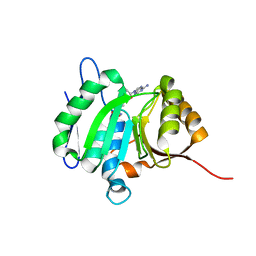 | |
8HL7
 
 | |
5EPP
 
 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
3NX2
 
 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
3QCA
 
 | |
3QC8
 
 | | Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Kim, K.H, Kang, W, Suh, S.W, Yang, J.K. | | Deposit date: | 2011-01-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Proteins, 79, 2011
|
|
