5STD
 
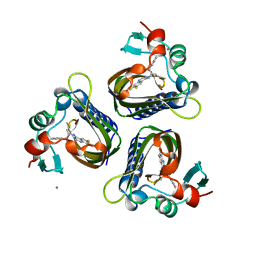 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 2 | | Descriptor: | (6,7-DIFLUORO-QUINAZOLIN-4-YL)-(1-METHYL-2,2-DIPHENYL-ETHYL)-AMINE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
6STD
 
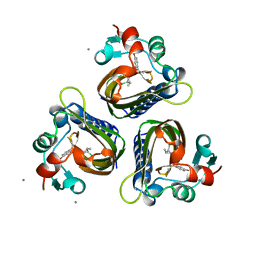 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 3 | | Descriptor: | 2,2-DICHLORO-1-METHANESULFINYL-3-METHYL-CYCLOPROPANECARBOXYLIC ACID [1-(4-BROMO-PHENYL)-ETHYL]-AMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
7STD
 
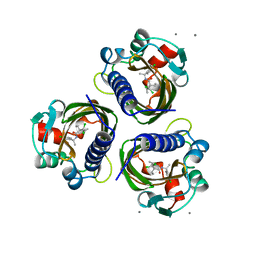 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4STD
 
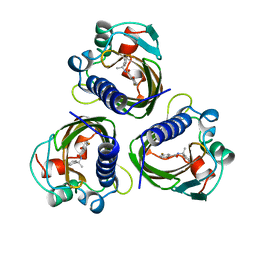 | | HIGH RESOLUTION STRUCTURES OF SCYTALONE DEHYDRATASE-INHIBITOR COMPLEXES CRYSTALLIZED AT PHYSIOLOGICAL PH | | Descriptor: | N-[1-(4-BROMOPHENYL)ETHYL]-5-FLUORO SALICYLAMIDE, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4EVM
 
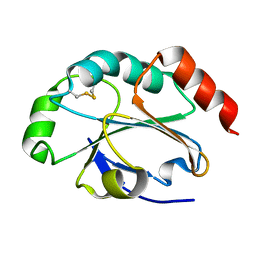 | | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A | | Descriptor: | Thioredoxin family protein | | Authors: | Wawrzak, Z, Stogios, P.J, Minasov, G, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A
To be Published
|
|
5AJR
 
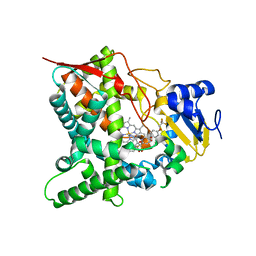 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with the 1-tetrazole derivative VT-1161 ((R)-2-(2,4-Difluorophenyl)-1, 1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl) pyridin-2-yl)propan-2-ol) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Wawrzak, Z, Hoekstra, W, I Lepesheva, G. | | Deposit date: | 2015-02-26 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Clinical Candidate Vt-1161'S Antiparasitic Effect in Vitro, Activity in a Murine Model of Chagas Disease, and Structural Characterization in Complex with the Target Enzyme Cyp51 from Trypanosoma Cruzi.
Antimicrob.Agents Chemother., 60, 2015
|
|
4H6O
 
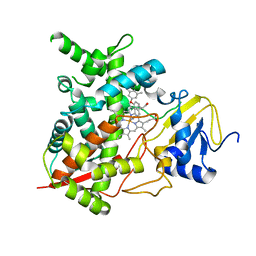 | | Sterol 14-alpha demethylase (CYP51)from Trypanosoma cruzi in complex with the inhibitor NEU321 (1-(3-(4-chloro-3,5-dimethylphenoxy)benzyl)-1H-imidazole | | Descriptor: | 1-[3-(4-chloro-3,5-dimethylphenoxy)benzyl]-1H-imidazole, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Wawrzak, Z, Hargrove, T.Y, Pollastri, M.P, Lepesheva, G.I. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antitrypanosomal Lead Discovery: Identification of a Ligand-Efficient Inhibitor of Trypanosoma cruzi CYP51 and Parasite Growth.
J.Med.Chem., 56, 2013
|
|
4LES
 
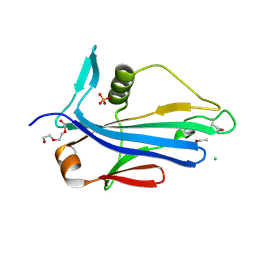 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
2A43
 
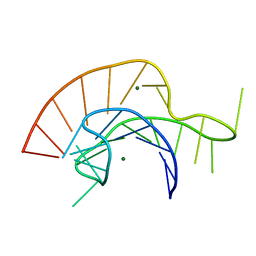 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
4CK8
 
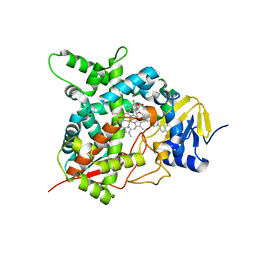 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CKA
 
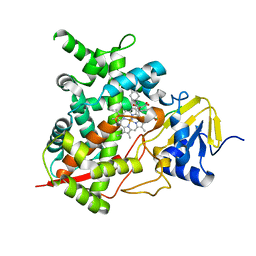 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFS) | | Descriptor: | (1S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-isopropylphenylcarbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CK9
 
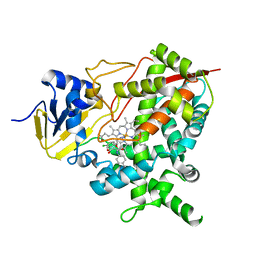 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
7RTQ
 
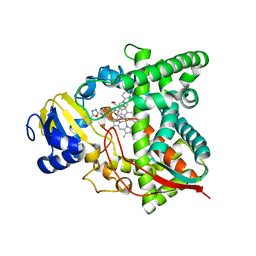 | | Sterol 14alpha demethylase (CYP51) from Naegleria fowleri in complex with an inhibitor R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | N-[(1R)-2-(1H-imidazol-1-yl)-1-(3,4',5-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, Protein CYP51 | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Relaxed Substrate Requirements of Sterol 14 alpha-Demethylase from Naegleria fowleri Are Accompanied by Resistance to Inhibition.
J.Med.Chem., 64, 2021
|
|
3R8Y
 
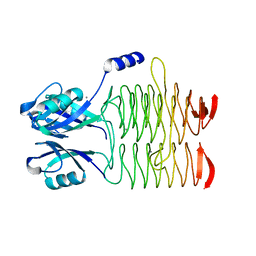 | | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, CALCIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase
TO BE PUBLISHED
|
|
3RUY
 
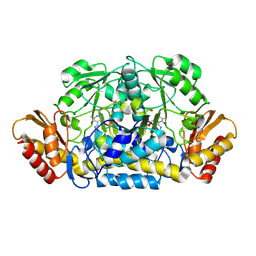 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | Descriptor: | Ornithine aminotransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
6FMO
 
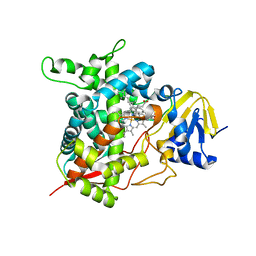 | |
6Q2T
 
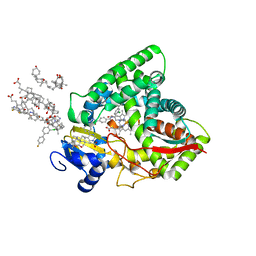 | | Human sterol 14a-demethylase (CYP51) in complex with the functionally irreversible inhibitor (R)-N-(1-(3-chloro-4'-fluoro-[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Lanosterol 14-alpha demethylase, N-[(1R)-1-(3-chloro-4'-fluoro[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, ... | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Validation of Human Sterol 14 alpha-Demethylase (CYP51) Druggability: Structure-Guided Design, Synthesis, and Evaluation of Stoichiometric, Functionally Irreversible Inhibitors.
J.Med.Chem., 62, 2019
|
|
2RF3
 
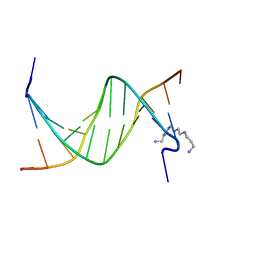 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
3KBO
 
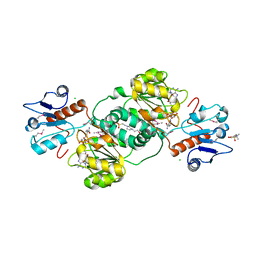 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
3L4D
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Leishmania infantum in complex with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-12-19 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate Preferences and Catalytic Parameters Determined by Structural Characteristics of Sterol 14{alpha}-Demethylase (CYP51) from Leishmania infantum.
J.Biol.Chem., 286, 2011
|
|
3L44
 
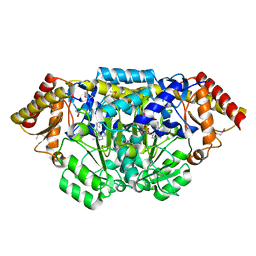 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
3KHM
 
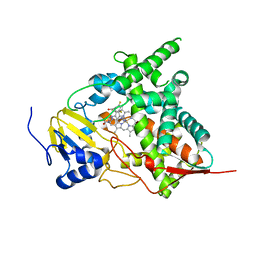 | | Crystal structure of sterol 14alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with inhibitor fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14 alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
2M11
 
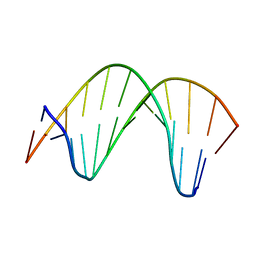 | | Structure of perimidinone-derived synthetic nucleoside paired with guanine in DNA duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(D3N)P*GP*CP*G)-3') | | Authors: | Kowal, E.A, Lad, R, Pallan, P.S, Muffly, E, Wawrzak, Z, Egli, M, Sturla, S.J, Stone, M.P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of O6-benzyl-2'-deoxyguanosine by a perimidinone-derived synthetic nucleoside: a DNA interstrand stacking interaction.
Nucleic Acids Res., 41, 2013
|
|
1TW7
 
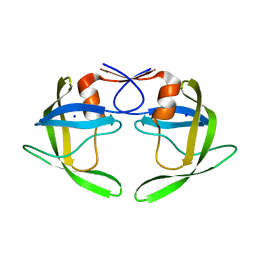 | | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target | | Descriptor: | SODIUM ION, protease | | Authors: | Martin, P, Vickrey, J.F, Proteasa, G, Jimenez, Y.L, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2004-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target
Structure, 13, 2005
|
|
1VRO
 
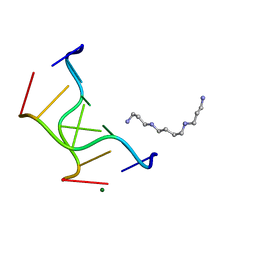 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
