5JPQ
 
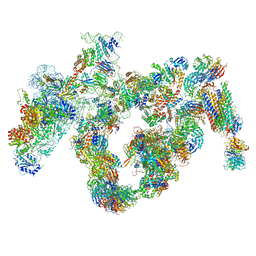 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
6H4N
 
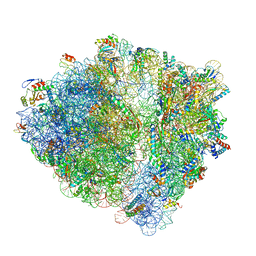 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
4RKD
 
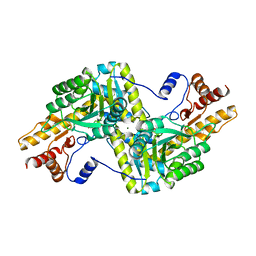 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with aspartic acid | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYLENE)-AMINO]-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aromatic amino acid aminotransferase, ... | | Authors: | Bujacz, A, Rutkiewicz-Krotewicz, M, Bujacz, G, Nowakowska-Sapota, K, Turkiewicz, M. | | Deposit date: | 2014-10-12 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure and enzymatic properties of a broad substrate-specificity psychrophilic aminotransferase from the Antarctic soil bacterium Psychrobacter sp. B6.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RKC
 
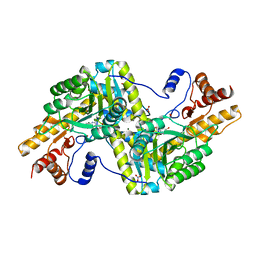 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aromatic amino acid aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rutkiewicz-Krotewicz, M, Bujacz, G, Nowakowska-Sapota, K, Turkiewicz, M. | | Deposit date: | 2014-10-12 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure and enzymatic properties of a broad substrate-specificity psychrophilic aminotransferase from the Antarctic soil bacterium Psychrobacter sp. B6.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1SEG
 
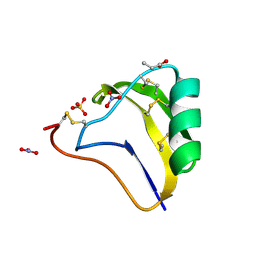 | | Crystal structure of a toxin chimera between Lqh-alpha-IT from the scorpion Leiurus quinquestriatus hebraeus and AAH2 from Androctonus australis hector | | Descriptor: | AAH2: LQH-ALPHA-IT (FACE) CHIMERIC TOXIN, NITRATE ION, PROPANOIC ACID, ... | | Authors: | Karbat, I, Frolow, F, Froy, O, Gilles, N, Cohen, L, Turkov, M, Gordon, D, Gurevitz, M. | | Deposit date: | 2004-02-17 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of the high insecticidal potency of scorpion alpha-toxins.
J.Biol.Chem., 279, 2004
|
|
2I61
 
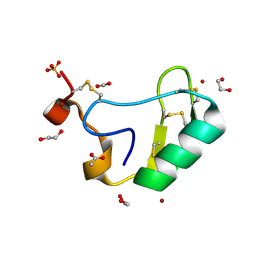 | | Depressant anti-insect neurotoxin, LqhIT2 from Leiurus quinquestriatus hebraeus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CARBON DIOXIDE, ... | | Authors: | Frolow, F, Gurevitz, M, Karbat, I, Turkov, M, Gordon, D. | | Deposit date: | 2006-08-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray Structure and Mutagenesis of the Scorpion Depressant Toxin LqhIT2 Reveals Key Determinants Crucial for Activity and Anti-Insect Selectivity.
J.Mol.Biol., 366, 2007
|
|
8SMC
 
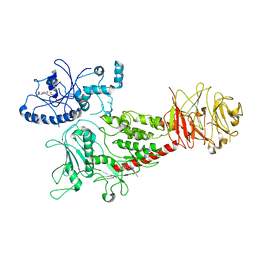 | | Cryo-EM structure of LRRK2 bound with type-I inhibitor DNL201 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, GUANOSINE-5'-DIPHOSPHATE, non-specific serine/threonine protein kinase | | Authors: | Sun, J, Zhu, H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2.
Science, 382, 2023
|
|
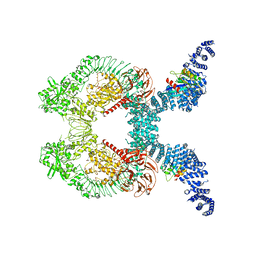
8FO2
 
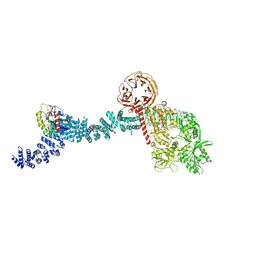 | |
8FO8
 
 | |
8FO9
 
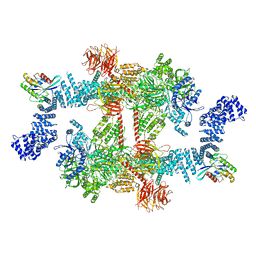 | |
2MLM
 
 | | Solution structure of sortase A from S. aureus in complex with benzo[d]isothiazol-3-one based inhibitor | | Descriptor: | N-{2-oxo-2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]ethyl}-2-sulfanylbenzamide, Sortase family protein | | Authors: | Jaudzems, K, Zhulenkovs, D, Leonchiks, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-11-19 | | Last modified: | 2018-06-27 | | Method: | SOLUTION NMR | | Cite: | Discovery and structure-activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase.
Bioorg.Med.Chem., 22, 2014
|
|
6ZUP
 
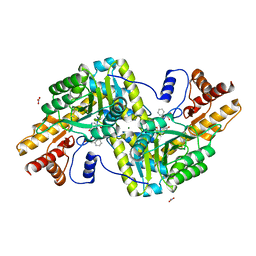 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
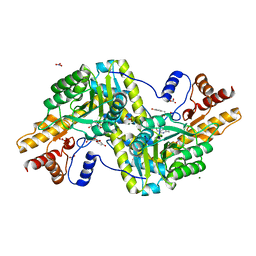 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZVG
 
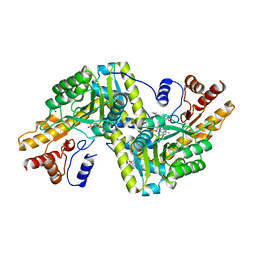 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-indole-3-lactic acid | | Descriptor: | 3-(INDOL-3-YL) LACTATE, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Rum, J, Rutkiewicz, M, Pruska, A, Bujacz, A, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6T3V
 
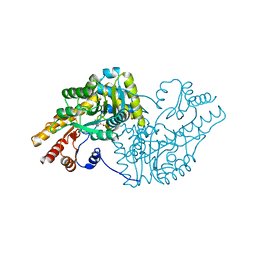 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
