2NTY
 
 | | Rop4-GDP-PRONE8 | | Descriptor: | Emb|CAB41934.1, GUANOSINE-5'-DIPHOSPHATE, Rac-like GTP-binding protein ARAC5 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
2NTX
 
 | | Prone8 | | Descriptor: | Emb|CAB41934.1 | | Authors: | Thomas, C, Fricke, I, Scrima, A, Berken, A, Wittinghofer, A. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Evidence for a Common Intermediate in Small G Protein-GEF Reactions
Mol.Cell, 25, 2007
|
|
3BWD
 
 | | Crystal structure of the plant Rho protein ROP5 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rac-like GTP-binding protein ARAC6 | | Authors: | Thomas, C, Berken, A. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of the plant Rho protein ROP5
To be Published
|
|
2WBL
 
 | | Three-dimensional structure of a binary ROP-PRONE complex | | Descriptor: | RAC-LIKE GTP-BINDING PROTEIN ARAC2, RHO OF PLANTS GUANINE NUCLEOTIDE EXCHANGE FACTOR 8 | | Authors: | Thomas, C, Fricke, I, Weyand, M, Berken, A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Biol.Chem., 390, 2009
|
|
3S9D
 
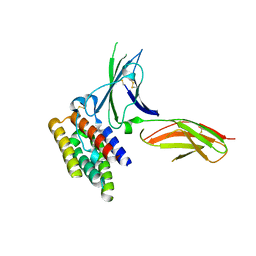 | | binary complex between IFNa2 and IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9999 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3SE3
 
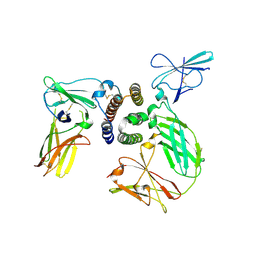 | | human IFNa2-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha 2b, Interferon alpha/beta receptor 1, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.0001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3SE4
 
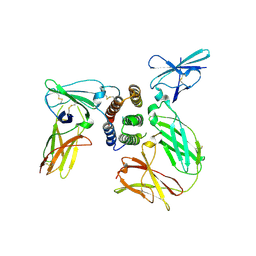 | | human IFNw-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha/beta receptor 1, Interferon alpha/beta receptor 2, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3S98
 
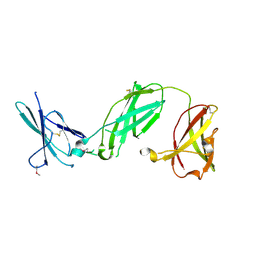 | | human IFNAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha/beta receptor 1 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3S8W
 
 | | D2 domain of human IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-05-31 | | Release date: | 2011-08-31 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
4DEP
 
 | | Structure of the IL-1b signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, Interleukin-1 receptor accessory protein, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2012-01-21 | | Release date: | 2012-03-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the activating IL-1 receptor signaling complex.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5OPI
 
 | | Crystal structure of the TAPBPR-MHC I peptide editing complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Thomas, C, Tampe, R. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing.
Science, 358, 2017
|
|
6RAM
 
 | | Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation with wider opened intracellular gate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAH
 
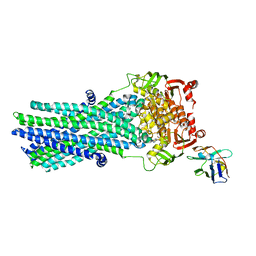 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAG
 
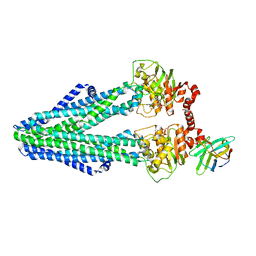 | | Heterodimeric ABC exporter TmrAB in inward-facing wide conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAN
 
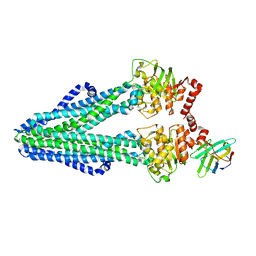 | | Heterodimeric ABC exporter TmrAB in inward-facing wide conformation | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, Nanobody Nb9F10 | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAJ
 
 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAK
 
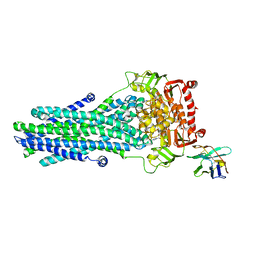 | | Heterodimeric ABC exporter TmrAB in vanadate trapped outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAI
 
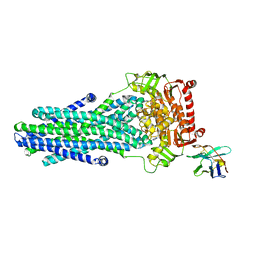 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAF
 
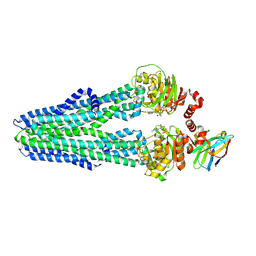 | | Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anti-vesicular stomatitis virus N VHH, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAL
 
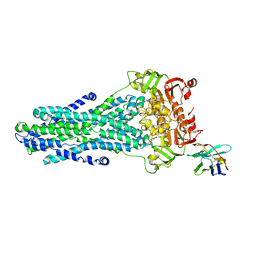 | | Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
7PHR
 
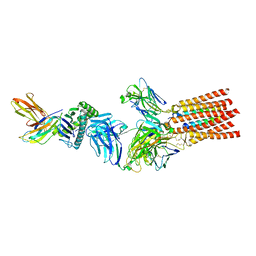 | | Structure of a fully assembled T-cell receptor engaging a tumor-associated peptide-MHC I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Susac, L, Thomas, C, Tampe, R. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC.
Cell, 185, 2022
|
|
7QPD
 
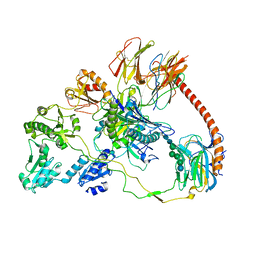 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
7QNG
 
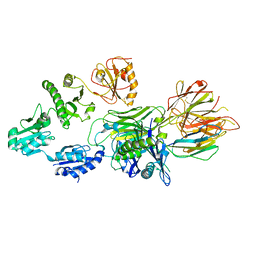 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
5IM5
 
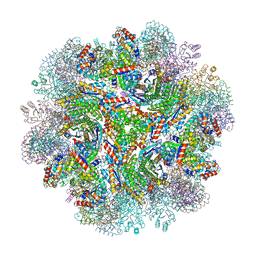 | | Crystal structure of designed two-component self-assembling icosahedral cage I53-40 | | Descriptor: | Designed Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase, Designed Riboflavin synthase | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.699 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
5IM6
 
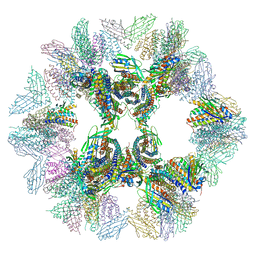 | | Crystal structure of designed two-component self-assembling icosahedral cage I32-28 | | Descriptor: | Designed self-assembling icosahedral cage I32-28 dimeric subunit, Designed self-assembling icosahedral cage I32-28 trimeric subunit | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.588 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
