1B23
 
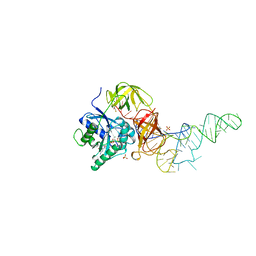 | | E. coli cysteinyl-tRNA and T. aquaticus elongation factor EF-TU:GTP ternary complex | | Descriptor: | CYSTEINE, CYSTEINYL TRNA, ELONGATION FACTOR TU, ... | | Authors: | Nissen, P, Kjeldgaard, M, Thirup, S, Nyborg, J. | | Deposit date: | 1998-12-04 | | Release date: | 1998-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA.
Structure Fold.Des., 7, 1999
|
|
1AYO
 
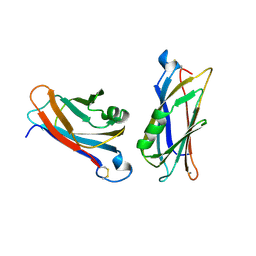 | | RECEPTOR BINDING DOMAIN OF BOVINE ALPHA-2-MACROGLOBULIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-2-MACROGLOBULIN, CALCIUM ION | | Authors: | Jenner, L.B, Husted, L, Thirup, S, Sottrup-Jensen, L, Nyborg, J. | | Deposit date: | 1997-11-07 | | Release date: | 1998-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the receptor-binding domain of alpha 2-macroglobulin.
Structure, 6, 1998
|
|
4XCM
 
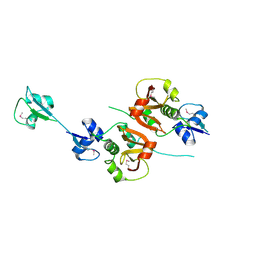 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1TUI
 
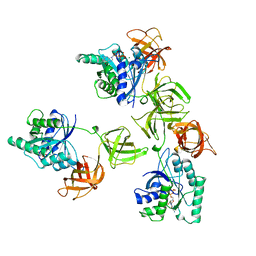 | | INTACT ELONGATION FACTOR TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Polekhina, G, Thirup, S, Kjeldgaard, M, Nissen, P, Lippmann, C, Nyborg, J. | | Deposit date: | 1996-05-23 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unwinding in the effector region of elongation factor EF-Tu-GDP.
Structure, 4, 1996
|
|
1TTT
 
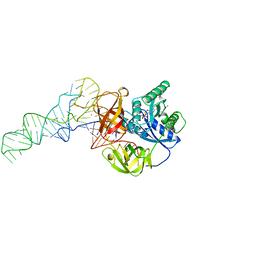 | | Phe-tRNA, elongation factoR EF-TU:GDPNP ternary complex | | Descriptor: | MAGNESIUM ION, OF ELONGATION FACTOR TU (EF-TU), PHENYLALANINE, ... | | Authors: | Nissen, P, Kjeldgaard, M, Thirup, S, Polekhina, G, Reshetnikova, L, Clark, B.F.C, Nyborg, J. | | Deposit date: | 1995-11-16 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog.
Science, 270, 1995
|
|
5MRI
 
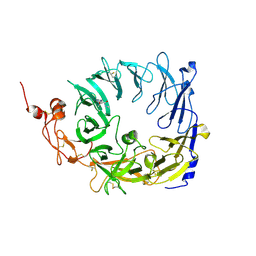 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5MRH
 
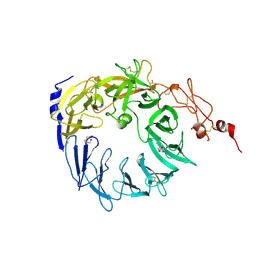 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-methylbutyl)-4~{H}-1,2,3-triazol-5-one, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1D2E
 
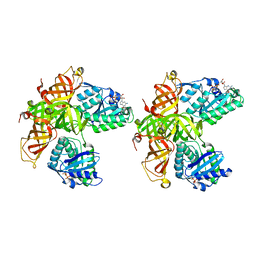 | | CRYSTAL STRUCTURE OF MITOCHONDRIAL EF-TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU (EF-TU), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Andersen, G.R, Thirup, S, Spremulli, L.L, Nyborg, J. | | Deposit date: | 1999-09-23 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP.
J.Mol.Biol., 297, 2000
|
|
4A1K
 
 | | Ykud L,D-transpeptidase | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4A1I
 
 | | ykud from B.subtilis | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE L, ... | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Three New Crystal Forms of the Ykud L,D-Transpeptidase from B. Subtilis.
To be Published
|
|
4A1J
 
 | | Ykud L,D-transpeptidase from B.subtilis | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Ykud
To be Published
|
|
1ETU
 
 | | STRUCTURAL DETAILS OF THE BINDING OF GUANOSINE DIPHOSPHATE TO ELONGATION FACTOR TU FROM E. COLI AS STUDIED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Clark, B.F.C, Lacour, T.F.M, Kjeldgaard, M, Morikawa, K, Nyborg, J, Rubin, R, Thirup, S. | | Deposit date: | 1988-01-15 | | Release date: | 1988-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography.
EMBO J., 4, 1985
|
|
1EFT
 
 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR EF-TU FROM THERMUS AQUATICUS IN THE GTP CONFORMATION | | Descriptor: | ELONGATION FACTOR TU, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kjeldgaard, M, Nissen, P, Thirup, S, Nyborg, J. | | Deposit date: | 1993-08-24 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation.
Structure, 1, 1993
|
|
4N7E
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF38469 | | Descriptor: | 2-[(6-methylpyridin-2-yl)carbamoyl]-5-(trifluoromethyl)benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The identification of AF38469: An orally bioavailable inhibitor of the VPS10P family sorting receptor Sortilin.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4MSL
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF40431 | | Descriptor: | N-[(7-hydroxy-4-methyl-2-oxo-2H-chromen-8-yl)methyl]-L-leucine, Sortilin, TETRAETHYLENE GLYCOL, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the first small-molecule ligand of the neuronal receptor sortilin and structure determination of the receptor-ligand complex.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
3KFU
 
 | | Crystal structure of the transamidosome | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C, ... | | Authors: | Blaise, M, Bailly, M, Frechin, M, Thirup, S, Becker, H.D, Kern, D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
Embo J., 29, 2010
|
|
5OCA
 
 | | PCSK9:Fab Complex with Dextran Sulfate | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-altropyranose-(1-6)-2,3-di-O-sulfo-alpha-L-glucopyranose, Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, ... | | Authors: | Thirup, S.S, Vilstrup, J.P. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan sulfate proteoglycans present PCSK9 to the LDL receptor.
Nat Commun, 8, 2017
|
|
4UZ3
 
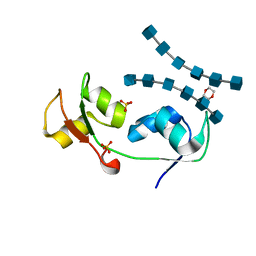 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1LU3
 
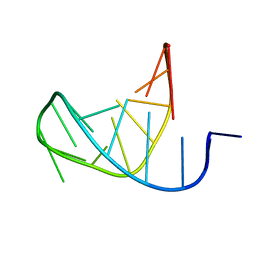 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | PHENYLALANINE TRANSFER RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1LS2
 
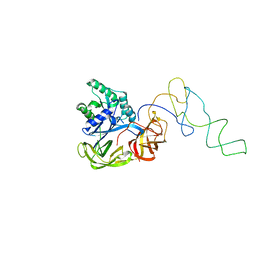 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | Elongation Factor Tu, Phenylalanine transfer RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-16 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
3F6K
 
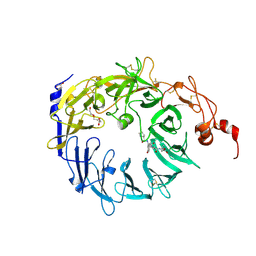 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with neurotensin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neurotensin, ... | | Authors: | Quistgaard, E.M, Madsen, P, Groftehauge, M.K, Nissen, P, Petersen, C.M, Thirup, S. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1EFC
 
 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
4UZ2
 
 | |
6TPT
 
 | | Crystal structures of FNIII domain three and four of the human leucocyte common antigen-related protein, LAR | | Descriptor: | Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
