4JHC
 
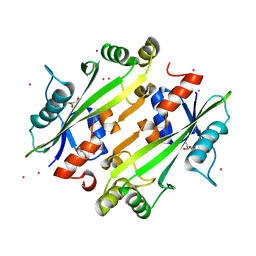 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli | | Descriptor: | GLYCEROL, Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2013-03-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4LU1
 
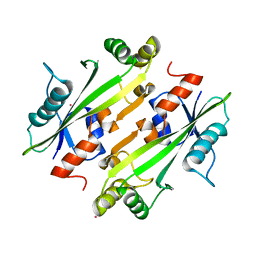 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli, mutant D69A | | Descriptor: | Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
5D8M
 
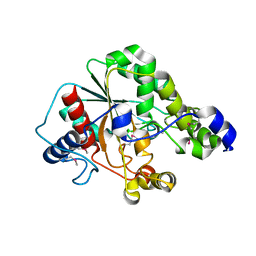 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | Descriptor: | Metagenomic carboxyl esterase MGS0156 | | Authors: | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-08-17 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of esterase (MGS0156)
To Be Published
|
|
4HEB
 
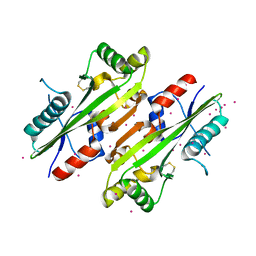 | | The Crystal structure of Maf protein of Bacillus subtilis | | Descriptor: | Septum formation protein Maf, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Brown, G, Flick, R, Tchigvintsev, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Iakounine, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
2P5X
 
 | | Crystal structure of Maf domain of human N-acetylserotonin O-methyltransferase-like protein | | Descriptor: | N-acetylserotonin O-methyltransferase-like protein, PHOSPHATE ION | | Authors: | Min, J, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
2R6O
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-18 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
3HHQ
 
 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3F4F
 
 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3Q3C
 
 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3P48
 
 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
1ZKI
 
 | | Structure of conserved protein PA5202 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, hypothetical protein PA5202 | | Authors: | Cuff, M.E, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3LMB
 
 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
4Q3M
 
 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
3VCR
 
 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | Descriptor: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3V77
 
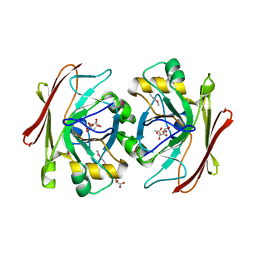 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | Descriptor: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3IRU
 
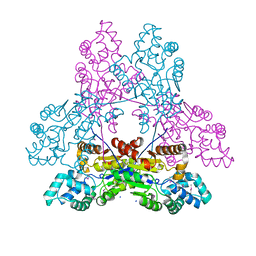 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | Descriptor: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
4Q3K
 
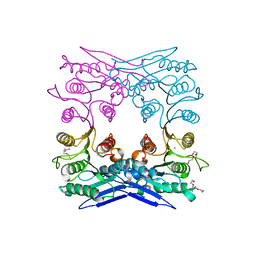 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3N
 
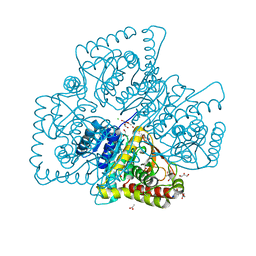 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3O
 
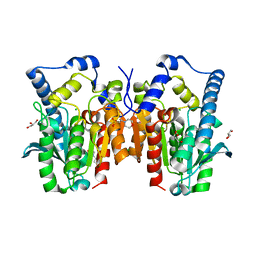 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
3S8Y
 
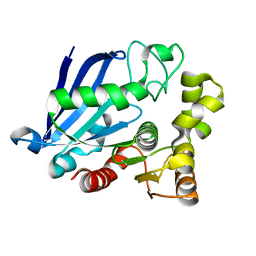 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
